+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13929 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 80S-bound human SKI complex in the open state | ||||||||||||||||||
 Map data Map data | postprocessed_map_hSKI_ribosome-bound_open | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | multiprotein complex / RNA helicase / DExH-box helicase / ATPase / RNA binding / RNA BINDING PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationSki complex / mRNA decay by 3' to 5' exoribonuclease / nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay / 3'-5' RNA helicase activity / Association of TriC/CCT with target proteins during biosynthesis / rescue of stalled ribosome / RNA helicase activity / RNA helicase / ATP hydrolysis activity / RNA binding ...Ski complex / mRNA decay by 3' to 5' exoribonuclease / nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay / 3'-5' RNA helicase activity / Association of TriC/CCT with target proteins during biosynthesis / rescue of stalled ribosome / RNA helicase activity / RNA helicase / ATP hydrolysis activity / RNA binding / ATP binding / nucleus / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
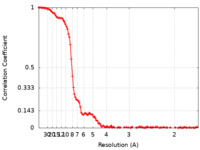
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | ||||||||||||||||||
 Authors Authors | Koegel A / Keidel A | ||||||||||||||||||
| Funding support |  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism. Authors: Alexander Kögel / Achim Keidel / Fabien Bonneau / Ingmar B Schäfer / Elena Conti /  Abstract: The superkiller (SKI) complex is the cytoplasmic co-factor and regulator of the RNA-degrading exosome. In human cells, the SKI complex functions mainly in co-translational surveillance-decay ...The superkiller (SKI) complex is the cytoplasmic co-factor and regulator of the RNA-degrading exosome. In human cells, the SKI complex functions mainly in co-translational surveillance-decay pathways, and its malfunction is linked to a severe congenital disorder, the trichohepatoenteric syndrome. To obtain insights into the molecular mechanisms regulating the human SKI (hSKI) complex, we structurally characterized several of its functional states in the context of 80S ribosomes and substrate RNA. In a prehydrolytic ATP form, the hSKI complex exhibits a closed conformation with an inherent gating system that effectively traps the 80S-bound RNA into the hSKI2 helicase subunit. When active, hSKI switches to an open conformation in which the gating is released and the RNA 3' end exits the helicase. The emerging picture is that the gatekeeping mechanism and architectural remodeling of hSKI underpin a regulated RNA channeling system that is mechanistically conserved among the cytoplasmic and nuclear helicase-exosome complexes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13929.map.gz emd_13929.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13929-v30.xml emd-13929-v30.xml emd-13929.xml emd-13929.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13929_fsc.xml emd_13929_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_13929.png emd_13929.png | 31.9 KB | ||
| Masks |  emd_13929_msk_1.map emd_13929_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13929.cif.gz emd-13929.cif.gz | 6.7 KB | ||
| Others |  emd_13929_additional_1.map.gz emd_13929_additional_1.map.gz emd_13929_half_map_1.map.gz emd_13929_half_map_1.map.gz emd_13929_half_map_2.map.gz emd_13929_half_map_2.map.gz | 50.1 MB 98.1 MB 98.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13929 http://ftp.pdbj.org/pub/emdb/structures/EMD-13929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13929 | HTTPS FTP |
-Validation report
| Summary document |  emd_13929_validation.pdf.gz emd_13929_validation.pdf.gz | 639 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13929_full_validation.pdf.gz emd_13929_full_validation.pdf.gz | 638.6 KB | Display | |
| Data in XML |  emd_13929_validation.xml.gz emd_13929_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_13929_validation.cif.gz emd_13929_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13929 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13929 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13929 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13929 | HTTPS FTP |
-Related structure data
| Related structure data |  7qe0MC  7qdrC  7qdsC  7qdyC  7qdzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13929.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13929.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed_map_hSKI_ribosome-bound_open | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
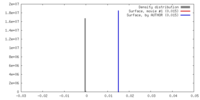
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13929_msk_1.map emd_13929_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
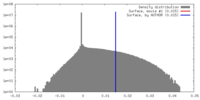
| Density Histograms |
-Additional map: postprocessed map hSKI-40S open
| File | emd_13929_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed_map_hSKI-40S_open | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_13929_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
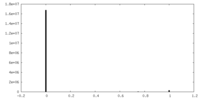
| Density Histograms |
-Half map: half map 2
| File | emd_13929_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
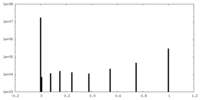
| Density Histograms |
- Sample components
Sample components
-Entire : H.s. SKI complex bound to the ribosome in open state
| Entire | Name: H.s. SKI complex bound to the ribosome in open state |
|---|---|
| Components |
|
-Supramolecule #1: H.s. SKI complex bound to the ribosome in open state
| Supramolecule | Name: H.s. SKI complex bound to the ribosome in open state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 380 KDa |
-Macromolecule #1: Helicase SKI2W
| Macromolecule | Name: Helicase SKI2W / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137.913688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMETERLVLP PPDPLDLPLR AVELGCTGHW ELLNLPGAPE SSLPHGLPPC APDLQQEAEQ LFLSSPAWLP LHGVEHSARK WQRKTDPWS LLAVLGAPVP SDLQAQRHPT TGQILGYKEV LLENTNLSAT TSLSLRRPPG PASQSLWGNP TQYPFWPGGM D EPTITDLN ...String: MMETERLVLP PPDPLDLPLR AVELGCTGHW ELLNLPGAPE SSLPHGLPPC APDLQQEAEQ LFLSSPAWLP LHGVEHSARK WQRKTDPWS LLAVLGAPVP SDLQAQRHPT TGQILGYKEV LLENTNLSAT TSLSLRRPPG PASQSLWGNP TQYPFWPGGM D EPTITDLN TREEAEEEID FEKDLLTIPP GFKKGMDFAP KDCPTPAPGL LSLSCMLEPL DLGGGDEDEN EAVGQPGGPR GD TVSASPC SAPLARASSL EDLVLKEAST AVSTPEAPEP PSQEQWAIPV DATSPVGDFY RLIPQPAFQW AFEPDVFQKQ AIL HLERHD SVFVAAHTSA GKTVVAEYAI ALAQKHMTRT IYTSPIKALS NQKFRDFRNT FGDVGLLTGD VQLHPEASCL IMTT EILRS MLYSGSDVIR DLEWVIFDEV HYINDVERGV VWEEVLIMLP DHVSIILLSA TVPNALEFAD WIGRLKRRQI YVIST VTRP VPLEHYLFTG NSSKTQGELF LLLDSRGAFH TKGYYAAVEA KKERMSKHAQ TFGAKQPTHQ GGPAQDRGVY LSLLAS LRT RAQLPVVVFT FSRGRCDEQA SGLTSLDLTT SSEKSEIHLF LQRCLARLRG SDRQLPQVLH MSELLNRGLG VHHSGIL PI LKEIVEMLFS RGLVKVLFAT ETFAMGVNMP ARTVVFDSMR KHDGSTFRDL LPGEYVQMAG RAGRRGLDPT GTVILLCK G RVPEMADLHR MMMGKPSQLQ SQFRLTYTMI LNLLRVDALR VEDMMKRSFS EFPSRKDSKA HEQALAELTK RLGALEEPD MTGQLVDLPE YYSWGEELTE TQHMIQRRIM ESVNGLKSLS AGRVVVVKNQ EHHNALGVIL QVSSNSTSRV FTTLVLCDKP LSQDPQDRG PATAEVPYPD DLVGFKLFLP EGPCDHTVVK LQPGDMAAIT TKVLRVNGEK ILEDFSKRQQ PKFKKDPPLA A VTTAVQEL LRLAQAHPAG PPTLDPVNDL QLKDMSVVEG GLRARKLEEL IQGAQCVHSP RFPAQYLKLR ERMQIQKEME RL RFLLSDQ SLLLLPEYHQ RVEVLRTLGY VDEAGTVKLA GRVACAMSSH ELLLTELMFD NALSTLRPEE IAALLSGLVC QSP GDAGDQ LPNTLKQGIE RVRAVAKRIG EVQVACGLNQ TVEEFVGELN FGLVEVVYEW ARGMPFSELA GLSGTPEGLV VRCI QRLAE MCRSLRGAAR LVGEPVLGAK METAATLLRR DIVFAASLYT Q UniProtKB: Superkiller complex protein 2 |
-Macromolecule #2: RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.710535 KDa |
| Sequence | String: UUUUUUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 6.0 sec. / Average electron dose: 67.62 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Target criteria: CORRELATION COEFFICIENT |
|---|---|
| Output model |  PDB-7qe0: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)