[English] 日本語
 Yorodumi
Yorodumi- EMDB-13590: Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13590 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 A: the structural basis for dimerisation | ||||||||||||
 Map data Map data | light harvesting core complex | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | light harvesting complex / photosynthesis / Cryo-EM / purple bacteria / RC-LH1 / RC-LH1-PufXYZ / dimer / dimeric core complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationplasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / : / photosynthetic electron transport in photosystem II / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Cereibacter sphaeroides 2.4.1 (bacteria) Cereibacter sphaeroides 2.4.1 (bacteria) | ||||||||||||
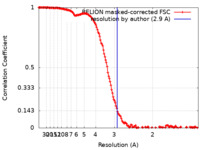
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Qian P / Hunter CN | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Biochem J / Year: 2021 Journal: Biochem J / Year: 2021Title: Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 Å: the structural basis for dimerisation. Authors: Pu Qian / Tristan I Croll / Andrew Hitchcock / Philip J Jackson / Jack H Salisbury / Pablo Castro-Hartmann / Kasim Sader / David J K Swainsbury / C Neil Hunter /   Abstract: The dimeric reaction centre light-harvesting 1 (RC-LH1) core complex of Rhodobacter sphaeroides converts absorbed light energy to a charge separation, and then it reduces a quinone electron and ...The dimeric reaction centre light-harvesting 1 (RC-LH1) core complex of Rhodobacter sphaeroides converts absorbed light energy to a charge separation, and then it reduces a quinone electron and proton acceptor to a quinol. The angle between the two monomers imposes a bent configuration on the dimer complex, which exerts a major influence on the curvature of the membrane vesicles, known as chromatophores, where the light-driven photosynthetic reactions take place. To investigate the dimerisation interface between two RC-LH1 monomers, we determined the cryogenic electron microscopy structure of the dimeric complex at 2.9 Å resolution. The structure shows that each monomer consists of a central RC partly enclosed by a 14-subunit LH1 ring held in an open state by PufX and protein-Y polypeptides, thus enabling quinones to enter and leave the complex. Two monomers are brought together through N-terminal interactions between PufX polypeptides on the cytoplasmic side of the complex, augmented by two novel transmembrane polypeptides, designated protein-Z, that bind to the outer faces of the two central LH1 β polypeptides. The precise fit at the dimer interface, enabled by PufX and protein-Z, by C-terminal interactions between opposing LH1 αβ subunits, and by a series of interactions with a bound sulfoquinovosyl diacylglycerol lipid, bring together each monomer creating an S-shaped array of 28 bacteriochlorophylls. The seamless join between the two sets of LH1 bacteriochlorophylls provides a path for excitation energy absorbed by one half of the complex to migrate across the dimer interface to the other half. #1:  Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2018 Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2018Title: Real-space refinement in PHENIX for cryo-EM and crystallography Authors: Qian P / Hunter CN | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13590.map.gz emd_13590.map.gz | 51.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13590-v30.xml emd-13590-v30.xml emd-13590.xml emd-13590.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13590_fsc.xml emd_13590_fsc.xml | 15.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13590.png emd_13590.png | 72.3 KB | ||
| Filedesc metadata |  emd-13590.cif.gz emd-13590.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13590 http://ftp.pdbj.org/pub/emdb/structures/EMD-13590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13590 | HTTPS FTP |
-Related structure data
| Related structure data |  7pqdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13590.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13590.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | light harvesting core complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Light harvesting core complex
+Supramolecule #1: Light harvesting core complex
+Macromolecule #1: LH1-alpha
+Macromolecule #2: LH1-beta
+Macromolecule #3: RC-H
+Macromolecule #4: RC-L
+Macromolecule #5: Reaction center protein M chain
+Macromolecule #6: PufZ
+Macromolecule #7: PufY
+Macromolecule #8: PufX
+Macromolecule #9: BACTERIOCHLOROPHYLL A
+Macromolecule #10: 3,4-DIHYDROSPHEROIDENE
+Macromolecule #11: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #12: (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(te...
+Macromolecule #13: UBIQUINONE-1
+Macromolecule #14: UBIQUINONE-10
+Macromolecule #15: BACTERIOPHEOPHYTIN A
+Macromolecule #16: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #17: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #18: FE (III) ION
+Macromolecule #19: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Component - Concentration: 20.0 mMol / Component - Formula: HEPES / Details: 20 mM HEPES, pH 7.8, 0.03% beta-DDM |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 277 K / Details: QF R1.2/1.3 300 mesh Cu grid. |
| Details | In 20 mM HEPES, pH 7.8, 0.03% beta-DDM buffer solution |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Max: 80.0 K |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 5058 / Average exposure time: 12.21 sec. / Average electron dose: 45.36 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)