[English] 日本語
 Yorodumi
Yorodumi- EMDB-11072: Reconstruction of bovine Ryanodine receptor 2 dimer of tetramers ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11072 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Reconstruction of bovine Ryanodine receptor 2 dimer of tetramers in complex with FKBP12.6 and nanobodies | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nanodisc / nanobody / FKBP12.6 / RyR2 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcyclic nucleotide binding / Stimuli-sensing channels / establishment of protein localization to endoplasmic reticulum / type B pancreatic cell apoptotic process / Purkinje myocyte to ventricular cardiac muscle cell signaling / regulation of atrial cardiac muscle cell action potential / Ion homeostasis / left ventricular cardiac muscle tissue morphogenesis / suramin binding / regulation of AV node cell action potential ...cyclic nucleotide binding / Stimuli-sensing channels / establishment of protein localization to endoplasmic reticulum / type B pancreatic cell apoptotic process / Purkinje myocyte to ventricular cardiac muscle cell signaling / regulation of atrial cardiac muscle cell action potential / Ion homeostasis / left ventricular cardiac muscle tissue morphogenesis / suramin binding / regulation of AV node cell action potential / regulation of SA node cell action potential / positive regulation of axon regeneration / regulation of ventricular cardiac muscle cell action potential / ventricular cardiac muscle cell action potential / : / embryonic heart tube morphogenesis / cardiac muscle hypertrophy / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / calcium ion transport into cytosol / negative regulation of release of sequestered calcium ion into cytosol / ryanodine-sensitive calcium-release channel activity / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to redox state / insulin secretion / protein peptidyl-prolyl isomerization / negative regulation of heart rate / cellular response to caffeine / response to vitamin E / FK506 binding / negative regulation of ryanodine-sensitive calcium-release channel activity / response to muscle activity / protein kinase A catalytic subunit binding / protein kinase A regulatory subunit binding / positive regulation of the force of heart contraction / smooth endoplasmic reticulum / regulation of ryanodine-sensitive calcium-release channel activity / smooth muscle contraction / detection of calcium ion / regulation of cytosolic calcium ion concentration / T cell proliferation / response to glucose / striated muscle contraction / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / positive regulation of heart rate / calcium channel inhibitor activity / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to muscle stretch / release of sequestered calcium ion into cytosol / cellular response to epinephrine stimulus / calcium channel complex / sarcoplasmic reticulum membrane / regulation of heart rate / sarcoplasmic reticulum / peptidylprolyl isomerase / establishment of localization in cell / peptidyl-prolyl cis-trans isomerase activity / response to hydrogen peroxide / sarcolemma / Z disc / intracellular calcium ion homeostasis / positive regulation of cytosolic calcium ion concentration / transmembrane transporter binding / response to hypoxia / calmodulin binding / intracellular membrane-bounded organelle / signaling receptor binding / calcium ion binding / identical protein binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
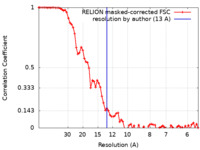
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Willegems K / Efremov R | |||||||||
| Funding support |  Belgium, 2 items Belgium, 2 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM. Authors: Chenyao Li / Katrien Willegems / Tomasz Uchański / Els Pardon / Jan Steyaert / Rouslan G Efremov /  Abstract: Ryanodine receptors (RyRs) are large Ca release channels residing in the endoplasmic or sarcoplasmic reticulum membrane. Three isoforms of RyRs have been identified in mammals, the disfunction of ...Ryanodine receptors (RyRs) are large Ca release channels residing in the endoplasmic or sarcoplasmic reticulum membrane. Three isoforms of RyRs have been identified in mammals, the disfunction of which has been associated with a series of life-threatening diseases. The need for large amounts of native tissue or eukaryotic cell cultures limits advances in structural studies of RyRs. Here, we report a method that utilizes nanobodies to purify RyRs from only 5 mg of total protein. The purification process, from isolated membranes to cryo-EM grade protein, is achieved within 4 h on the bench, yielding protein usable for cryo-EM analysis. This is demonstrated by solving the structures of rabbit RyR1, solubilized in detergent, reconstituted into lipid nanodiscs or liposomes, and bovine RyR2 reconstituted in nanodisc, and mouse RyR2 in detergent. The reported method facilitates structural studies of RyRs directed toward drug development and is useful in cases where the amount of starting material is limited. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11072.map.gz emd_11072.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11072-v30.xml emd-11072-v30.xml emd-11072.xml emd-11072.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11072_fsc.xml emd_11072_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11072.png emd_11072.png | 84.9 KB | ||
| Masks |  emd_11072_msk_1.map emd_11072_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11072.cif.gz emd-11072.cif.gz | 4.8 KB | ||
| Others |  emd_11072_half_map_1.map.gz emd_11072_half_map_1.map.gz emd_11072_half_map_2.map.gz emd_11072_half_map_2.map.gz | 20.3 MB 20.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11072 http://ftp.pdbj.org/pub/emdb/structures/EMD-11072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11072 | HTTPS FTP |
-Related structure data
| Related structure data |  8rrsC  8rrtC  8rruC  8rrvC  8rrwC  8rrxC  8rs0C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11072.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11072.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11072_msk_1.map emd_11072_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_11072_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_11072_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of bovine Ryanodine receptor 2
| Entire | Name: Dimer of bovine Ryanodine receptor 2 |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of bovine Ryanodine receptor 2
| Supramolecule | Name: Dimer of bovine Ryanodine receptor 2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Complex of Ryanodine receptor 2 with auxiliary protein FKBP12.6 and high affinity nanobody |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.2 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: GRAPHENE OXIDE / Support film - #1 - topology: CONTINUOUS | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293.15 K / Instrument: GATAN CRYOPLUNGE 3 Details: .Grid was blotted manually from the back in CP3 cryoplunge (Gatan) for 2s. | ||||||||||||||||||
| Details | The RyR2 tetrameric channels formed channel-dimers with the nanobody at the dimer-interface bound to the repeat12 domain of the individual channels |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 4088 / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)