[English] 日本語
 Yorodumi
Yorodumi- PDB-8rrx: Structure of RyR1 reconstituted into lipid nanodisc in primed sta... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8rrx | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of RyR1 reconstituted into lipid nanodisc in primed state in complex with Ca2+, ATP, caffeine and Nb9657 | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / Ion channel / Ca2+ / tetramer | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationATP-gated ion channel activity / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / cellular response to caffeine / skin development / organelle membrane / intracellularly gated calcium channel activity ...ATP-gated ion channel activity / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / cellular response to caffeine / skin development / organelle membrane / intracellularly gated calcium channel activity / smooth endoplasmic reticulum / outflow tract morphogenesis / regulation of ryanodine-sensitive calcium-release channel activity / toxic substance binding / striated muscle contraction / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / voltage-gated calcium channel activity / skeletal muscle fiber development / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / release of sequestered calcium ion into cytosol / sarcoplasmic reticulum membrane / cellular response to calcium ion / muscle contraction / sarcoplasmic reticulum / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / sarcolemma / calcium ion transmembrane transport / calcium channel activity / Z disc / intracellular calcium ion homeostasis / disordered domain specific binding / protein homotetramerization / transmembrane transporter binding / calmodulin binding / intracellular membrane-bounded organelle / calcium ion binding / ATP binding / identical protein binding / membrane Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Li, C. / Efremov, R.G. | ||||||||||||
| Funding support |  Belgium, European Union, 3items Belgium, European Union, 3items
| ||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM. Authors: Chenyao Li / Katrien Willegems / Tomasz Uchański / Els Pardon / Jan Steyaert / Rouslan G Efremov /  Abstract: Ryanodine receptors (RyRs) are large Ca release channels residing in the endoplasmic or sarcoplasmic reticulum membrane. Three isoforms of RyRs have been identified in mammals, the disfunction of ...Ryanodine receptors (RyRs) are large Ca release channels residing in the endoplasmic or sarcoplasmic reticulum membrane. Three isoforms of RyRs have been identified in mammals, the disfunction of which has been associated with a series of life-threatening diseases. The need for large amounts of native tissue or eukaryotic cell cultures limits advances in structural studies of RyRs. Here, we report a method that utilizes nanobodies to purify RyRs from only 5 mg of total protein. The purification process, from isolated membranes to cryo-EM grade protein, is achieved within 4 h on the bench, yielding protein usable for cryo-EM analysis. This is demonstrated by solving the structures of rabbit RyR1, solubilized in detergent, reconstituted into lipid nanodiscs or liposomes, and bovine RyR2 reconstituted in nanodisc, and mouse RyR2 in detergent. The reported method facilitates structural studies of RyRs directed toward drug development and is useful in cases where the amount of starting material is limited. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8rrx.cif.gz 8rrx.cif.gz | 3.5 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8rrx.ent.gz pdb8rrx.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8rrx.json.gz 8rrx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rr/8rrx https://data.pdbj.org/pub/pdb/validation_reports/rr/8rrx ftp://data.pdbj.org/pub/pdb/validation_reports/rr/8rrx ftp://data.pdbj.org/pub/pdb/validation_reports/rr/8rrx | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  19468MC  8rrsC  8rrtC  8rruC  8rrvC  8rrwC  8rs0C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 8 molecules HLJFIADG
| #1: Protein | Mass: 11667.305 Da / Num. of mol.: 4 / Source method: isolated from a natural source / Source: (natural)  #2: Protein | Mass: 564931.750 Da / Num. of mol.: 4 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Antibody , 1 types, 4 molecules KCEM
| #3: Antibody | Mass: 15125.495 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 20 molecules 


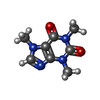





| #4: Chemical | ChemComp-ZN / #5: Chemical | ChemComp-POV / ( #6: Chemical | ChemComp-ATP / #7: Chemical | ChemComp-CFF / #8: Chemical | ChemComp-CA / |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||||||
| Buffer solution | pH: 7.4 | ||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: JEOL CRYO ARM 300 |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 1500 nm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: JEOL CRYOSPECPORTER |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Image processing | Details: The movies were selected after MotionCor correction and CTF refinement with the parameters: total drift less than 30 angstrom and resolution better than 5 angstrom | ||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2233299 Details: Particles were picked by template picking in Cryosparc | ||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C4 (4 fold cyclic) | ||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 175535 / Algorithm: FOURIER SPACE / Symmetry type: POINT |
 Movie
Movie Controller
Controller
























 PDBj
PDBj








