[English] 日本語
 Yorodumi
Yorodumi- EMDB-11071: Structure of Ryanodine Receptor 2 in lipid nano discs in complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11071 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Ryanodine Receptor 2 in lipid nano discs in complex with nanobody and FKBP12.6 in presence of calcium | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nanodisc / nanobody / FKBP12.6 / RyR2 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcyclic nucleotide binding / Stimuli-sensing channels / establishment of protein localization to endoplasmic reticulum / type B pancreatic cell apoptotic process / Purkinje myocyte to ventricular cardiac muscle cell signaling / regulation of atrial cardiac muscle cell action potential / Ion homeostasis / left ventricular cardiac muscle tissue morphogenesis / suramin binding / regulation of AV node cell action potential ...cyclic nucleotide binding / Stimuli-sensing channels / establishment of protein localization to endoplasmic reticulum / type B pancreatic cell apoptotic process / Purkinje myocyte to ventricular cardiac muscle cell signaling / regulation of atrial cardiac muscle cell action potential / Ion homeostasis / left ventricular cardiac muscle tissue morphogenesis / suramin binding / regulation of AV node cell action potential / regulation of SA node cell action potential / positive regulation of axon regeneration / regulation of ventricular cardiac muscle cell action potential / ventricular cardiac muscle cell action potential / : / embryonic heart tube morphogenesis / cardiac muscle hypertrophy / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / calcium ion transport into cytosol / negative regulation of release of sequestered calcium ion into cytosol / ryanodine-sensitive calcium-release channel activity / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to redox state / insulin secretion / protein peptidyl-prolyl isomerization / negative regulation of heart rate / cellular response to caffeine / response to vitamin E / FK506 binding / negative regulation of ryanodine-sensitive calcium-release channel activity / response to muscle activity / protein kinase A catalytic subunit binding / protein kinase A regulatory subunit binding / positive regulation of the force of heart contraction / smooth endoplasmic reticulum / regulation of ryanodine-sensitive calcium-release channel activity / smooth muscle contraction / detection of calcium ion / regulation of cytosolic calcium ion concentration / T cell proliferation / response to glucose / striated muscle contraction / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / positive regulation of heart rate / calcium channel inhibitor activity / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to muscle stretch / release of sequestered calcium ion into cytosol / cellular response to epinephrine stimulus / calcium channel complex / sarcoplasmic reticulum membrane / regulation of heart rate / sarcoplasmic reticulum / peptidylprolyl isomerase / establishment of localization in cell / peptidyl-prolyl cis-trans isomerase activity / response to hydrogen peroxide / sarcolemma / Z disc / intracellular calcium ion homeostasis / positive regulation of cytosolic calcium ion concentration / transmembrane transporter binding / response to hypoxia / calmodulin binding / intracellular membrane-bounded organelle / signaling receptor binding / calcium ion binding / identical protein binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | |||||||||
 Authors Authors | Willegems K / Efremov R | |||||||||
| Funding support |  Belgium, 2 items Belgium, 2 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM. Authors: Chenyao Li / Katrien Willegems / Tomasz Uchański / Els Pardon / Jan Steyaert / Rouslan G Efremov /  Abstract: Ryanodine receptors (RyRs) are large Ca release channels residing in the endoplasmic or sarcoplasmic reticulum membrane. Three isoforms of RyRs have been identified in mammals, the disfunction of ...Ryanodine receptors (RyRs) are large Ca release channels residing in the endoplasmic or sarcoplasmic reticulum membrane. Three isoforms of RyRs have been identified in mammals, the disfunction of which has been associated with a series of life-threatening diseases. The need for large amounts of native tissue or eukaryotic cell cultures limits advances in structural studies of RyRs. Here, we report a method that utilizes nanobodies to purify RyRs from only 5 mg of total protein. The purification process, from isolated membranes to cryo-EM grade protein, is achieved within 4 h on the bench, yielding protein usable for cryo-EM analysis. This is demonstrated by solving the structures of rabbit RyR1, solubilized in detergent, reconstituted into lipid nanodiscs or liposomes, and bovine RyR2 reconstituted in nanodisc, and mouse RyR2 in detergent. The reported method facilitates structural studies of RyRs directed toward drug development and is useful in cases where the amount of starting material is limited. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11071.map.gz emd_11071.map.gz | 167.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11071-v30.xml emd-11071-v30.xml emd-11071.xml emd-11071.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11071_fsc.xml emd_11071_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_11071.png emd_11071.png | 131.3 KB | ||
| Filedesc metadata |  emd-11071.cif.gz emd-11071.cif.gz | 8.6 KB | ||
| Others |  emd_11071_half_map_1.map.gz emd_11071_half_map_1.map.gz emd_11071_half_map_2.map.gz emd_11071_half_map_2.map.gz | 163.9 MB 163.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11071 http://ftp.pdbj.org/pub/emdb/structures/EMD-11071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11071 | HTTPS FTP |
-Related structure data
| Related structure data |  8rrsC  8rrtC  8rruC  8rrvC  8rrwC  8rrxC  8rs0C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11071.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11071.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_11071_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
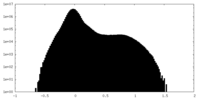
| Density Histograms |
-Half map: #1
| File | emd_11071_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
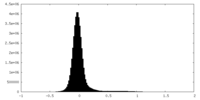
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Ryanodine receptor 2 with auxiliary protein FKBP12.6 a...
| Entire | Name: Complex of Ryanodine receptor 2 with auxiliary protein FKBP12.6 and high affinity nanobody |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Ryanodine receptor 2 with auxiliary protein FKBP12.6 a...
| Supramolecule | Name: Complex of Ryanodine receptor 2 with auxiliary protein FKBP12.6 and high affinity nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.2 MDa |
-Macromolecule #1: Ryanodine receptor 2
| Macromolecule | Name: Ryanodine receptor 2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: SNLSGSGEKT DDEVVLQCTA TIHKEQQKLC LAAEGFGNRL CFLESTSNSK NVPPDLSICT FVLEQSLSV RALQEMLANT VEKSEGAKEN PCTAQGGGHR TLLYGHAILL RHSYSGMYLC C LSTSRSST DKLAFDVGLQ EDTTGEACWW IIHPASKQRS EGEKVRVGDD ...String: SNLSGSGEKT DDEVVLQCTA TIHKEQQKLC LAAEGFGNRL CFLESTSNSK NVPPDLSICT FVLEQSLSV RALQEMLANT VEKSEGAKEN PCTAQGGGHR TLLYGHAILL RHSYSGMYLC C LSTSRSST DKLAFDVGLQ EDTTGEACWW IIHPASKQRS EGEKVRVGDD LILVSVSSER YL HLSYGNG SLHVDAAFQQ TLWSVAPISS GSEAAQGYLI GGDVLRLLHG HMDECLTVPS GEH GEEQRR TVHYEGGAVS VHARSLWRLE TLRVAWSGSH IRWGQPFRLR HVTTGKYLSL MEDK SLLLM DKEKADVKST AFTFRSSKEK LDVGVRKEVD GMGTSEIKYG DSVCYIQHIN TGLWL TYQS VDVKSVRMGS IQRKAIMHHE GHMDDGLNLS RSQHEESRTA RVIRSTVFLF NRFIRG LDA LSKKVKASTV DLPIESVSLS LQDLIGYFHP PDEHLEHEDK QNRLRALKNR QNLFQEE GM INLVLECIDR LHVYSSAAHF ADVAGREAGE SWKSILNSLY ELLAALIRGN RKNCAQFS G SLDWLISRLE RLEASSGILE VLHCVLVESP EALNIIKEGH IKSIISLLDK HGRNHKVLD VLCSLCVCHG VAVRSNQHLI CDNLLPGRDL LLQTRLVNHV SSMRPNIFLG VSEGSAQYKK WYYELMVDH TEPFVTAEAT HLRVGWASTE GYSPYPGGGE EWGGNGVGDD LFSYGFDGLH L WSGCIART VSSPNQHLLR TDDVISCCLD LSAPSISFRI NGQPVQGMFE NFNIDGLFFP VV SFSAGIK VRFLLGGRHG EFKFLPPPGY APCYEAVLPK EKLKVEHSRE YKQERTYTRD LLG PTVSLT QAAFTPIPVD TSQIVLPPHL ERIREKLAEN IHELWVMNKI ELGWQYGPVR DDNK RQHPC LVEFSKLPEQ ERNYNLQMSL ETLKTLLALG CHVGISDEHA EEKVKKMKLP KNYQL TSGY KPAPMDLSFI KLTPSQEAMV DKLAENAHNV WARDRIRQGW TYGIQQDVKN RRNPRL VPY ALLDDRTKKS NKDSLREAVR TLLGYGYNLE APDQDHAARA EVCSGTGERF RIFRAEK TY AVKAGRWYFE FEAVTAGDMR VGWCRPGCQP DQELGSDERA FAFDGFKAQR WHQGNEHY G RSWQAGDVVG CMVDMTEHTM MFTLNGEILL DDSGSELAFK DFDVGDGFIP VCSLGVAQV GRLNFGKDVS TLKYFTICGL QEGYEPFAVN TNRDITMWLS KRLPQFLQVP SNHEHIEVTR IDGTIDSSP CLKVTQKSFG SQNSSTDIMF YRLSMPIECA EVFSKTAAGG IPGTGLFGPK N DLEDYDVD SDFEVLMKTA HGHLVPDRGD RDKEATKPEF NNHKDYAQEK PSRLKQRFLL RR TKPDYST SHSARLTEDV LADERDDYDY LMQTSTYYYS VRIFPGQEPA NVWVGWITSD FHQ YDTGFD LDRVRTVTVT LGDEKGKVHE SIKRSNCYMV CAGESLSPGQ GRNNNGLEIG CVVD AASGL LTFTANGKEL STYYQVEPST KLFPAVFAQA TSPNVFQFEL GRIKNVMPLS AGLFK SEHK NPVPQCPPRL HVQFLSHVLW SRMPNQFLKV DVSRISERQG WLVQCMEPLQ FMSLHI PEE NRSVDILELT EQEELLKFHY HTLRLYSAVC ALGNHRVAHA LCSHVDEPQL LYAIENK YM PGLLRTGYYD LLIDIHLSSY ATARLMMNNE FIVPMTEETK SITLFPDEKK KHGLPGIG L STSLRPRMQF SSPSFVSINT EGYQYSPEFP LDILKAKTIQ MLTEAVKEGS LHARDPVGG TTEFLFVPLI KLFYTLLIMG VFHNEDLKHV LQLIEPSVFK EAASPEEESE VAEKEPFVED SKLEGAAEE ENKGAKRPKE GLLQMKLPEP VKLQMCLLLQ YLCDCQVRHR IEAIVAFSDD F VAKLQDNQ RFRYNEVMQA LNMSAALTAR KTKEFRSPPQ EQINMLLNFK DDKSECPCPE EI RDQLLDF HEDLMTHCGI ELDEDRSLDG NNDLTIRGRL LSLVEKVTYL KKKQAEKPVE SDS KKSSTL QQLISETMVR WAQESVIEDP ELVRAMFVLL HRQYDGIGGL VRALPKTYTI NGVS VEDTI NLLASLGQIR SLLSVRMGKE EEKLMIRGLG DIMNNKVFYQ HPNLMRALGM HETVM EVMV NVLGGGESKE ITFPKMVANC CRFLCYFCRI SRQNQKAMFD HLSYLLENSS VGLASP AMR GSTPLDVAAA SVMDNNELAL ALREPDLEKV VRYLAGCGLQ SCQMLVSKGY PDIGWNP VE GERYLDFLRF AVFCNGESVE ENANVVVRLL IRRPECFGPA LRGEGGNGLL AAMEEAIK I AEDPSRDGPS PTTGSSKTPD TEEEEDDTIH MGNAIMTFYS ALIDLLGRCA PEMHLIHAA KGEAIRIRSI LRSLIPLGDL VGVISIAFQM PTIAKDGNVV EPDMSAGFCP DHKAAMVLFL DRVYGIEVQ DFLLHLLEVG FLPDLRAAAS LDTAALSATD MALALNRYLC TAVLPLLTRC A PLFAGTEH HASLIDSLLH TVYRLSKGCS LTKAQRDSIE VCLLSICGQL RPSMMQHLLR RL VFDVPLL NEHAKMPLKL LTNHYERCWK YYCLPGGWGN FGAASEEELH LSRKLFWGIF DAL SQKKYE QELFKLALPC LSAVAGALPP DYMESNYVSM MEKQSSMDSE GNFNPQPVDT SNIT IPEKL EYFINKYAEH AHDKWSMDKL ANGWIYGEIY SDSSKVQPLM KPYKLLSEKE KEIYR WPIK ESLKTMLAWG WRIERTREGD SMALYNRTRR ISQTSQVSVD AAHGYSPRAI DMSNVT LSR DLHAMAEMMA ENYHNIWAKK KKLELEAKGG GNHPLLVPYD TLTAKEKAKD REKAQDI LK FLQINGYAVS RGFKDLELDT PSIEKRFAYS FLQQLIRYVD EAHQYILEFD GGSRSKGE H FPYEQEIKFF AKVVLPLIDQ YFKNHRLYFL SAASRPLCSG GHASNKEKEM NFSEQFIQL FVSSLGNDAT SIVNCLHILG QTLDARTVMK TGLESVKSAL RAFLDNAAED LEKTMENLKQ GQFTHTRNQ PKGVTQIINY TTVALLPMLS SLFEHIGQHQ FGEDLILEDV QVSCYRILTS L YALGTSKS IYVERQRSAL GECLAAFAGA FPVAFLETHL DKHNIYSIYN TKSSRERAAL NL PANVEDV CPNIPSLEKL MEEIVDLAES GIRYTQMPHV MEVVLPMLCS YMSRWWEHGP ENN PGRAEM CCTALNSEHM NTLLGNILKI IYNNLGIDEG AWMKRLAVFS QPIINKVKPQ LLKT HFLPL MEKLKKKAAM VVSEEDHLKS EARGDMSEAE LLILDEFTTL ARDLYAFYPL LIRFV DYNR AKWLKEPNQE AEDLFRMVAE VFIYWSKSHN FKREEQNFVV QNEINNMSFL IMDTKS KMS KAAVSDQERK KMKRKGDRYS MQTSLIVAAL KRLLPIGLNI CAPGDQELIA LAKNRFS LK DTEDEVRDII RSNIHLQGKL EDPAIRWQMA LYKDLPNRTE DTSDPEKTVE RVLDIANV L FHLEQVSFCV EHPQRSKKAV WHKLLSKQRK RAVVACFRMA PLYNLPRHRA VNLFLQGYE KSWIETEEHY FEDKLIEDLA KPGAEPPEED EGTKRVDPLH QLILLFSRTA LTEKWYGWGS CHDEEDDDG EEEVKSFEEK EMEKQKLLYQ QARLHDRGAA EMVLQTISAS KGETGPMVAA T LKLGIAIL NGGNSTVQQK MLDYLKEKKD VGFFQSLAGL MQSCSVLDLN AFERQNKAEG LG MVTEEGS GEKVLQDDEF TCDLFRFLQL LCEGHNSVQN WTDVIDEQGQ RNFSKAIQVA KQV FNTLTE YIQGPCTGNQ QSLAHSRLWD AVVGFLHVFA HMQMKLSQDS SQIELLKELM DLQK DMVVM LLSMLEGNVV NGTIGKQMVD MLVESSNNVE MILKFFDMFL KLKDLTSSDT FKEYD PDGK GVISKRDFHK AMESHKHYTQ SETEFLLSCA ETDENETLDY EEFVKRFHEP AKDIGF NVA VLLTNLSEHM PNDTRLQTFL ELAESVLNYF QPFLGRIEIM GSAKRIERVY FEISESS RT QWEKPQVKES KRQFIFDVVN EGGEKEKMEL FVNFCEDTIF EMQLAAQISE SDLNERSA N KEESEKEKPE EQGPRMGFFS IMTVKSALFA LRYNILTLMR MLSLKSLKKQ MKKVKKMTV KDMITAFFTS YWSILMSLLH FVASVFRGFS RIIGSLLLGG SLVEGAKKIK VAELLANMPD PTQDEVRGD GEEGERKTLE GALPSEDLTD LKELSEESDL LSDIFGLDLK REGGQYKLIP H NPNAGLSD LMSNPVPIPE VQEKFQEQKA KEEEKEEKEE SKSEPEKAEG EDGEKEEKAK ED KGKQKLR QLHTHRYGEP EVPESAFWKK IIAYQQKLLN YFARNFYNMR MLALFVAFAI NFI LLFYKV STSSVVEGKE LPTRSSSENA RVSSLDSSSP RIIAVHYVLE ESSGYMEPTL RILA ILHTV ISFFCIIGYY CLKVPLVIFK REKEVARKLE FDGLYITEQP SEDDIKGQWD RLVIN TQSF PNNYWDKFVK RKVMDKYGEF YGRDRISELL GMDKAALDFS DAREKKKPKK DSSLSA VLN SIDVKYQMWK LGVVFTDNSF LYLAWYMTMS ILGHYNNFFF AAHLLDIAMG FKTLRTI LS SVTHNGKQLV LTVGLLAVVV YLYTVVAFNF FRKFYNKSED GDTPDMKCDD MLTCYMFH M YVGVRAGGGI GDEIEDPAGD EYEIYRIIFD ITFFFFVIVI LLAIIQGLII DAFGELRDQ QEQVKEDMET KCFICGIGND YFDTVPHGFE THTLQEHNLA NYLFFLMYLI NKDETEHTGQ ESYVWKMYQ ERCWEFFPAG DCFRKQYEDQ LN UniProtKB: UNIPROTKB: E1BE29 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: GRAPHENE OXIDE / Support film - #1 - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 400.0 kPa Details: Grid was glow discharged before the application of fresh graphene oxide, 30min prior to plunging | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293.15 K / Instrument: GATAN CRYOPLUNGE 3 Details: .Grid was blotted manually from the back in CP3 cryoplunge (Gatan) for 2s. | ||||||||||||||||||
| Details | The RyR2 tetrameric channels formed channel-dimers with the nanobody at the dimer-interface bound to the repeat12 domain of the individual channels |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 4088 / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)