[English] 日本語
 Yorodumi
Yorodumi- EMDB-11053: Cryo-EM density map of nanobody-4C8 bound, human CD9 in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11053 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM density map of nanobody-4C8 bound, human CD9 in complex with EWI-F-deltaIg1-5 | |||||||||
 Map data Map data | Local-resolution filtered, masked cryo-EM density map of the EWI-F%u0394Ig1-5 - CD9 - 4C8 complex, sharpened with B = -1200 A2. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationAcrosome Reaction and Sperm:Oocyte Membrane Binding / myoblast fusion involved in skeletal muscle regeneration / sperm-egg recognition / fusion of sperm to egg plasma membrane involved in single fertilization / regulation of macrophage migration / paranodal junction assembly / glial cell migration / platelet alpha granule membrane / negative regulation of platelet aggregation / oligodendrocyte development ...Acrosome Reaction and Sperm:Oocyte Membrane Binding / myoblast fusion involved in skeletal muscle regeneration / sperm-egg recognition / fusion of sperm to egg plasma membrane involved in single fertilization / regulation of macrophage migration / paranodal junction assembly / glial cell migration / platelet alpha granule membrane / negative regulation of platelet aggregation / oligodendrocyte development / Uptake and function of diphtheria toxin / cellular response to low-density lipoprotein particle stimulus / clathrin-coated endocytic vesicle membrane / platelet activation / receptor internalization / integrin binding / endocytic vesicle membrane / Platelet degranulation / extracellular vesicle / cell population proliferation / cell adhesion / apical plasma membrane / negative regulation of cell population proliferation / external side of plasma membrane / focal adhesion / protein-containing complex / extracellular space / extracellular exosome / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.6 Å | |||||||||
 Authors Authors | Oosterheert W / Gros P | |||||||||
| Funding support |  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2020 Journal: Life Sci Alliance / Year: 2020Title: Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F. Authors: Wout Oosterheert / Katerina T Xenaki / Viviana Neviani / Wouter Pos / Sofia Doulkeridou / Jip Manshande / Nicholas M Pearce / Loes Mj Kroon-Batenburg / Martin Lutz / Paul Mp van Bergen En ...Authors: Wout Oosterheert / Katerina T Xenaki / Viviana Neviani / Wouter Pos / Sofia Doulkeridou / Jip Manshande / Nicholas M Pearce / Loes Mj Kroon-Batenburg / Martin Lutz / Paul Mp van Bergen En Henegouwen / Piet Gros /  Abstract: Tetraspanins are eukaryotic membrane proteins that contribute to a variety of signaling processes by organizing partner-receptor molecules in the plasma membrane. How tetraspanins bind and cluster ...Tetraspanins are eukaryotic membrane proteins that contribute to a variety of signaling processes by organizing partner-receptor molecules in the plasma membrane. How tetraspanins bind and cluster partner receptors into tetraspanin-enriched microdomains is unknown. Here, we present crystal structures of the large extracellular loop of CD9 bound to nanobodies 4C8 and 4E8 and, the cryo-EM structure of 4C8-bound CD9 in complex with its partner EWI-F. CD9-EWI-F displays a tetrameric arrangement with two central EWI-F molecules, dimerized through their ectodomains, and two CD9 molecules, one bound to each EWI-F transmembrane helix through CD9-helices h3 and h4. In the crystal structures, nanobodies 4C8 and 4E8 bind CD9 at loops C and D, which is in agreement with the 4C8 conformation in the CD9-EWI-F complex. The complex varies from nearly twofold symmetric (with the two CD9 copies nearly anti-parallel) to ca. 50° bent arrangements. This flexible arrangement of CD9-EWI-F with potential CD9 homo-dimerization at either end provides a "concatenation model" for forming short linear or circular assemblies, which may explain the occurrence of tetraspanin-enriched microdomains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11053.map.gz emd_11053.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11053-v30.xml emd-11053-v30.xml emd-11053.xml emd-11053.xml | 40.7 KB 40.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11053_fsc.xml emd_11053_fsc.xml | 3.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11053.png emd_11053.png | 56.9 KB | ||
| Masks |  emd_11053_msk_1.map emd_11053_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Others |  emd_11053_additional_1.map.gz emd_11053_additional_1.map.gz emd_11053_additional_2.map.gz emd_11053_additional_2.map.gz emd_11053_additional_3.map.gz emd_11053_additional_3.map.gz emd_11053_additional_4.map.gz emd_11053_additional_4.map.gz emd_11053_additional_5.map.gz emd_11053_additional_5.map.gz emd_11053_additional_6.map.gz emd_11053_additional_6.map.gz emd_11053_additional_7.map.gz emd_11053_additional_7.map.gz emd_11053_half_map_1.map.gz emd_11053_half_map_1.map.gz emd_11053_half_map_2.map.gz emd_11053_half_map_2.map.gz | 2.7 MB 2.4 MB 411 KB 1.1 MB 1.1 MB 1.1 MB 1.1 MB 2.8 MB 2.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11053 http://ftp.pdbj.org/pub/emdb/structures/EMD-11053 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11053 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11053 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11053.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11053.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local-resolution filtered, masked cryo-EM density map of the EWI-F%u0394Ig1-5 - CD9 - 4C8 complex, sharpened with B = -1200 A2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.0855 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
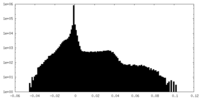
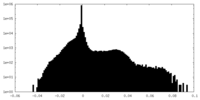
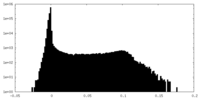
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Additional map: Refined, unsharpened cryo-EM density map of the EWI-F%u0394Ig1-5...
+Additional map: Local-resolution filtered, masked cryo-EM density map of the...
+Additional map: Unsharpened cryo-EM density map of the EWI-F%u0394Ig1-5 -...
+Additional map: Class #1 of a 3D classification of the...
+Additional map: Class #2 of a 3D classification of the...
+Additional map: Class #3 of a 3D classification of the...
+Additional map: Class #4 of a 3D classification of the...
+Half map: Unfiltered half map 1 the EWI-F%u0394Ig1-5 - CD9 - 4C8 complex.
+Half map: Unfiltered half map 2 the EWI-F%u0394Ig1-5 - CD9 - 4C8 complex.
- Sample components
Sample components
-Entire : Nanobody-4C8 bound, human CD9 in complex with EWI-F-deltaIg1-5
| Entire | Name: Nanobody-4C8 bound, human CD9 in complex with EWI-F-deltaIg1-5 |
|---|---|
| Components |
|
-Supramolecule #1: Nanobody-4C8 bound, human CD9 in complex with EWI-F-deltaIg1-5
| Supramolecule | Name: Nanobody-4C8 bound, human CD9 in complex with EWI-F-deltaIg1-5 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The complex is arranged as a hetero-hexamer: 4C8 - CD9 - EWI-F - EWI-F - CD9 - 4C8 |
|---|---|
| Molecular weight | Theoretical: 188 KDa |
-Supramolecule #2: CD9 - EWI-F-deltaIg1-5 complex
| Supramolecule | Name: CD9 - EWI-F-deltaIg1-5 complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 Details: The complex was purified from HEK293 GNT1- cells as a hetero-tetramer, arranged as CD9 - EWI-F - EWI-F - CD9. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 GNT1- Homo sapiens (human) / Recombinant cell: HEK293 GNT1- |
| Molecular weight | Theoretical: 158 KDa |
-Supramolecule #3: Nanobody 4C8 molecule 2
| Supramolecule | Name: Nanobody 4C8 molecule 2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 Details: The nanobody was purified from a bacterial expression system. |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 30 KDa |
-Macromolecule #1: CD9
| Macromolecule | Name: CD9 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSWSHPQFEK GSWSHPQFEK GSPVKGGTKC IKYLLFGFNF IFWLAGIAVL AIGLWLRFDS QTKSIFEQET NNNNSSFYTG VYILIGAGAL MMLVGFLGCC GAVQESQCML GLFFGFLLVI FAIEIAAAIW GYSHKDEVIK EVQEFYKDTY NKLKTKDEPQ RETLKAIHYA ...String: GSWSHPQFEK GSWSHPQFEK GSPVKGGTKC IKYLLFGFNF IFWLAGIAVL AIGLWLRFDS QTKSIFEQET NNNNSSFYTG VYILIGAGAL MMLVGFLGCC GAVQESQCML GLFFGFLLVI FAIEIAAAIW GYSHKDEVIK EVQEFYKDTY NKLKTKDEPQ RETLKAIHYA LNCCGLAGGV EQFISDICPK KDVLETFTVK SCPDAIKEVF DNKFHIIGAV GIGIAVVMIF GMIFSMILCC AIRRNREMVA AA |
-Macromolecule #2: EWI-F / CD9-partner-1 / FPRP
| Macromolecule | Name: EWI-F / CD9-partner-1 / FPRP / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGSTSGPIFN ASVHSDTPSV IRGDLIKLFC IITVEGAALD PDDMAFDVSW FAVHSFGLDK APVLLSSLDR KGIVTTSRRD WKSDLSLERV SVLEFLLQVH GSEDQDFGNY YCSVTPWVKS PTGSWQKEAE IHSKPVFITV KMDVLNAFKY PLLIGVGLST VIGLLSCLIG ...String: MGSTSGPIFN ASVHSDTPSV IRGDLIKLFC IITVEGAALD PDDMAFDVSW FAVHSFGLDK APVLLSSLDR KGIVTTSRRD WKSDLSLERV SVLEFLLQVH GSEDQDFGNY YCSVTPWVKS PTGSWQKEAE IHSKPVFITV KMDVLNAFKY PLLIGVGLST VIGLLSCLIG YCSSHWCCKK EVQETRRERR RLMSMEMDAA AHHHHHH |
-Macromolecule #3: Nanobody 4C8
| Macromolecule | Name: Nanobody 4C8 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQAGGSLRL SCAASGRTFS DYVMGWFRQA PGKERTFVAR IGWSGDLTYY ADSVKGRFTI SRDNAKNTVY LQMNSLKPED TAIYYCAADE RWGTGGKFDY WGQGTQVTVS SHGSGLVPRG SGGGHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
Details: 50 mM Tris pH 7.8, 150 mM NaCl, 0.08 % (w/v) digitonin. | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV Details: The blotting was performed at 20 degrees Celsius for four seconds with blot force 1.. | ||||||||||||
| Details | The CD9 - EWI-F-deltaIg1-5 was purified from HEK293 GNT1- cells. Nanobody 4C8 was added to the thew complex prior to size-exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-36 / Number grids imaged: 1 / Number real images: 10724 / Average exposure time: 7.2 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Details | Models were rigid-body fitted into the map using UCSF Chimera and not refined. |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)