[English] 日本語
 Yorodumi
Yorodumi- EMDB-0406: Single-particle cryo-EM reconstruction of horse liver alcohol deh... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0406 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-particle cryo-EM reconstruction of horse liver alcohol dehydrogenase using 200 keV | |||||||||
 Map data Map data | Single-particle cryo-EM reconstruction of horse liver alcohol dehydrogenase (sharpened) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dehydrogenase / NADH-binding / homo-2-mer / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationall-trans-retinol dehydrogenase (NAD+) activity / alcohol dehydrogenase / retinoic acid metabolic process / retinol metabolic process / zinc ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
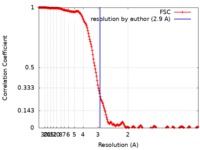
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Herzik Jr MA / Wu M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: High-resolution structure determination of sub-100 kDa complexes using conventional cryo-EM. Authors: Mark A Herzik / Mengyu Wu / Gabriel C Lander /  Abstract: Determining high-resolution structures of biological macromolecules amassing less than 100 kilodaltons (kDa) has been a longstanding goal of the cryo-electron microscopy (cryo-EM) community. While ...Determining high-resolution structures of biological macromolecules amassing less than 100 kilodaltons (kDa) has been a longstanding goal of the cryo-electron microscopy (cryo-EM) community. While the Volta phase plate has enabled visualization of specimens in this size range, this instrumentation is not yet fully automated and can present technical challenges. Here, we show that conventional defocus-based cryo-EM methodologies can be used to determine high-resolution structures of specimens amassing less than 100 kDa using a transmission electron microscope operating at 200 keV coupled with a direct electron detector. Our ~2.7 Å structure of alcohol dehydrogenase (82 kDa) proves that bound ligands can be resolved with high fidelity to enable investigation of drug-target interactions. Our ~2.8 Å and ~3.2 Å structures of methemoglobin demonstrate that distinct conformational states can be identified within a dataset for proteins as small as 64 kDa. Furthermore, we provide the sub-nanometer cryo-EM structure of a sub-50 kDa protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0406.map.gz emd_0406.map.gz | 176.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0406-v30.xml emd-0406-v30.xml emd-0406.xml emd-0406.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0406_fsc.xml emd_0406_fsc.xml | 21.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0406.png emd_0406.png | 155.6 KB | ||
| Masks |  emd_0406_msk_1.map emd_0406_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0406.cif.gz emd-0406.cif.gz | 6.9 KB | ||
| Others |  emd_0406_additional.map.gz emd_0406_additional.map.gz emd_0406_half_map_1.map.gz emd_0406_half_map_1.map.gz emd_0406_half_map_2.map.gz emd_0406_half_map_2.map.gz | 172.4 MB 408.2 MB 408.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0406 http://ftp.pdbj.org/pub/emdb/structures/EMD-0406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0406 | HTTPS FTP |
-Related structure data
| Related structure data |  6nbbMC  0407C  0408C  0409C  6nbcC  6nbdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10249 (Title: Horse liver alcohol dehydrogenase movies obtained using Talos Arctica operating at 200 kV equipped with a K2 EMPIAR-10249 (Title: Horse liver alcohol dehydrogenase movies obtained using Talos Arctica operating at 200 kV equipped with a K2Data size: 1.3 TB Data #1: Raw, unaligned movies of horse liver alcohol dehydrogenase acquired on a Talos Arctica using a K2 direct electron detector [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0406.map.gz / Format: CCP4 / Size: 190.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0406.map.gz / Format: CCP4 / Size: 190.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM reconstruction of horse liver alcohol dehydrogenase (sharpened) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.5585 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
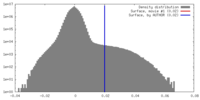
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0406_msk_1.map emd_0406_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
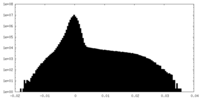
| Density Histograms |
-Additional map: Single-particle cryo-EM reconstruction of horse liver alcohol dehydrogenase...
| File | emd_0406_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM reconstruction of horse liver alcohol dehydrogenase (unsharpened) | ||||||||||||
| Projections & Slices |
| ||||||||||||
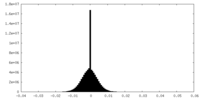
| Density Histograms |
-Half map: Even half map
| File | emd_0406_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
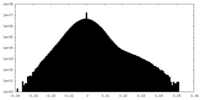
| Density Histograms |
-Half map: Odd half map
| File | emd_0406_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Odd half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
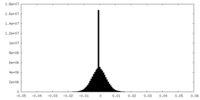
| Density Histograms |
- Sample components
Sample components
-Entire : Alcohol dehydrogenase from equine liver
| Entire | Name: Alcohol dehydrogenase from equine liver |
|---|---|
| Components |
|
-Supramolecule #1: Alcohol dehydrogenase from equine liver
| Supramolecule | Name: Alcohol dehydrogenase from equine liver / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Lyophilized horse liver ADH purchased from Sigma Aldrich was further purified to homogeneity. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81 KDa |
-Macromolecule #1: Alcohol dehydrogenase E chain
| Macromolecule | Name: Alcohol dehydrogenase E chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: alcohol dehydrogenase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.853273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: STAGKVIKCK AAVLWEEKKP FSIEEVEVAP PKAHEVRIKM VATGICRSDD HVVSGTLVTP LPVIAGHEAA GIVESIGEGV TTVRPGDKV IPLFTPQCGK CRVCKHPEGN FCLKNDLSMP RGTMQDGTSR FTCRGKPIHH FLGTSTFSQY TVVDEISVAK I DAASPLEK ...String: STAGKVIKCK AAVLWEEKKP FSIEEVEVAP PKAHEVRIKM VATGICRSDD HVVSGTLVTP LPVIAGHEAA GIVESIGEGV TTVRPGDKV IPLFTPQCGK CRVCKHPEGN FCLKNDLSMP RGTMQDGTSR FTCRGKPIHH FLGTSTFSQY TVVDEISVAK I DAASPLEK VCLIGCGFST GYGSAVKVAK VTQGSTCAVF GLGGVGLSVI MGCKAAGAAR IIGVDINKDK FAKAKEVGAT EC VNPQDYK KPIQEVLTEM SNGGVDFSFE VIGRLDTMVT ALSCCQEAYG VSVIVGVPPD SQNLSMNPML LLSGRTWKGA IFG GFKSKD SVPKLVADFM AKKFALDPLI THVLPFEKIN EGFDLLRSGE SIRTILTF UniProtKB: Alcohol dehydrogenase E chain |
-Macromolecule #2: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 2 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 6 sec. / Pretreatment - Atmosphere: OTHER Details: Grids were plasma cleaned using a Solarus plasma cleaner (Gatan, Inc.). | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER Details: Sample was manually blotted for 4-5 seconds using Whatman No. 1 filter paper immediately prior to plunge-freezing.. | |||||||||||||||
| Details | Lyophilized horse liver ADH (Sigma Aldrich) was solubilized and further purified to homogeneity. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-44 / Number grids imaged: 2 / Number real images: 1151 / Average exposure time: 11.0 sec. / Average electron dose: 69.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 16.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 73000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)