[English] 日本語
 Yorodumi
Yorodumi- EMDB-0232: Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosom... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0232 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the body of the small mitoribosomal subunit in complex with mt-IF-3 | |||||||||
 Map data Map data | map of the T. brucei mitoribosome small subunit body in complex with mt-IF3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitoribosome / translation / Trypanosoma / small ribosomal subunit / 9S rRNA / ribosomal protein / mitochondrial initiation factor IF-3 / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationmodulation of formation of structure involved in a symbiotic process / organellar small ribosomal subunit / mitochondrial mRNA editing complex / mitochondrial RNA processing / quorum sensing / kinetoplast / nuclear lumen / ciliary plasm / mitochondrial small ribosomal subunit / mRNA stabilization ...modulation of formation of structure involved in a symbiotic process / organellar small ribosomal subunit / mitochondrial mRNA editing complex / mitochondrial RNA processing / quorum sensing / kinetoplast / nuclear lumen / ciliary plasm / mitochondrial small ribosomal subunit / mRNA stabilization / superoxide dismutase / fatty acid beta-oxidation / superoxide dismutase activity / protein kinase A regulatory subunit binding / axoneme / structural constituent of ribosome / ribosome / translation / mitochondrion / RNA binding / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Ramrath DJF / Niemann M | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Authors: David J F Ramrath / Moritz Niemann / Marc Leibundgut / Philipp Bieri / Céline Prange / Elke K Horn / Alexander Leitner / Daniel Boehringer / André Schneider / Nenad Ban /  Abstract: Ribosomal RNA (rRNA) plays key functional and architectural roles in ribosomes. Using electron microscopy, we determined the atomic structure of a highly divergent ribosome found in mitochondria of , ...Ribosomal RNA (rRNA) plays key functional and architectural roles in ribosomes. Using electron microscopy, we determined the atomic structure of a highly divergent ribosome found in mitochondria of , a unicellular parasite that causes sleeping sickness in humans. The trypanosomal mitoribosome features the smallest rRNAs and contains more proteins than all known ribosomes. The structure shows how the proteins have taken over the role of architectural scaffold from the rRNA: They form an autonomous outer shell that surrounds the entire particle and stabilizes and positions the functionally important regions of the rRNA. Our results also reveal the "minimal" set of conserved rRNA and protein components shared by all ribosomes that help us define the most essential functional elements. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0232.map.gz emd_0232.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0232-v30.xml emd-0232-v30.xml emd-0232.xml emd-0232.xml | 63.9 KB 63.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0232.png emd_0232.png | 50.8 KB | ||
| Filedesc metadata |  emd-0232.cif.gz emd-0232.cif.gz | 16.5 KB | ||
| Others |  emd_0232_half_map_1.map.gz emd_0232_half_map_1.map.gz emd_0232_half_map_2.map.gz emd_0232_half_map_2.map.gz | 98.4 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0232 http://ftp.pdbj.org/pub/emdb/structures/EMD-0232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0232 | HTTPS FTP |
-Related structure data
| Related structure data |  6hiyMC  0229C  0230C  0231C  0233C  6hivC  6hiwC  6hixC  6hizC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0232.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0232.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of the T. brucei mitoribosome small subunit body in complex with mt-IF3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
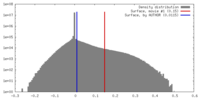
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map (even) of the T. brucei mitoribosome...
| File | emd_0232_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map (even) of the T. brucei mitoribosome small subunit body in complex with mt-IF3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map (odd) of the T. brucei mitoribosome...
| File | emd_0232_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map (odd) of the T. brucei mitoribosome small subunit body in complex with mt-IF3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : body of the T. brucei mitoribosome small subunit in complex with ...
+Supramolecule #1: body of the T. brucei mitoribosome small subunit in complex with ...
+Macromolecule #1: mS48
+Macromolecule #2: mS51
+Macromolecule #3: mS56
+Macromolecule #4: mS59
+Macromolecule #5: mS60
+Macromolecule #6: mS61
+Macromolecule #7: mS62
+Macromolecule #8: mS63
+Macromolecule #9: mS64
+Macromolecule #10: mS65
+Macromolecule #11: mS66
+Macromolecule #12: mS68
+Macromolecule #13: mS73
+Macromolecule #14: mS74
+Macromolecule #15: mS55m
+Macromolecule #16: bS6m
+Macromolecule #17: uS8m
+Macromolecule #18: uS9m
+Macromolecule #19: uS11m
+Macromolecule #20: uS12m
+Macromolecule #21: uS15m
+Macromolecule #22: bS16m
+Macromolecule #23: uS17m
+Macromolecule #24: bS18m
+Macromolecule #25: uS21m
+Macromolecule #26: mt-IF-3
+Macromolecule #27: mS22
+Macromolecule #28: mS23
+Macromolecule #29: mS26
+Macromolecule #30: mS34
+Macromolecule #31: mS37
+Macromolecule #32: mS38
+Macromolecule #33: mS41
+Macromolecule #34: mS42
+Macromolecule #35: mS43
+Macromolecule #36: mS47
+Macromolecule #38: Unknown protein
+Macromolecule #39: Unknown protein
+Macromolecule #40: Unknown protein
+Macromolecule #41: Unknown protein
+Macromolecule #37: 9S rRNA
+Macromolecule #42: ZINC ION
+Macromolecule #43: MAGNESIUM ION
+Macromolecule #44: SPERMIDINE
+Macromolecule #45: SPERMINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 98 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | We used Coot and O for initial model building and refined the structure using Phenix |
|---|---|
| Refinement | Protocol: AB INITIO MODEL / Overall B value: 41.4 |
| Output model |  PDB-6hiy: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)