+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3364 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-tomogram averaging of Tula virus glycoprotein spike | |||||||||
 Map data Map data | Sub-tomogram average of Tula virus surface | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tula virus / membrane protein / glycoprotein / hantavirus / bunyavirus / receptor binding | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host TRAF-mediated signal transduction / host cell Golgi membrane / host cell mitochondrion / host cell surface / host cell endoplasmic reticulum membrane / symbiont-mediated suppression of host innate immune response / : / virion membrane / cell surface / signal transduction ...symbiont-mediated suppression of host TRAF-mediated signal transduction / host cell Golgi membrane / host cell mitochondrion / host cell surface / host cell endoplasmic reticulum membrane / symbiont-mediated suppression of host innate immune response / : / virion membrane / cell surface / signal transduction / membrane / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Tula virus Tula virus | |||||||||
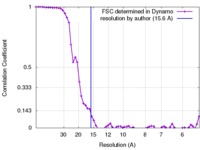
| Method | subtomogram averaging / cryo EM / Resolution: 15.6 Å | |||||||||
 Authors Authors | Li S / Rissanen I / Zeltina A / Hepojoki J / Raghwani J / Harlos K / Pybus OG / Huiskonen JT / Bowden TA | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2016 Journal: Cell Rep / Year: 2016Title: A Molecular-Level Account of the Antigenic Hantaviral Surface. Authors: Sai Li / Ilona Rissanen / Antra Zeltina / Jussi Hepojoki / Jayna Raghwani / Karl Harlos / Oliver G Pybus / Juha T Huiskonen / Thomas A Bowden /   Abstract: Hantaviruses, a geographically diverse group of zoonotic pathogens, initiate cell infection through the concerted action of Gn and Gc viral surface glycoproteins. Here, we describe the high- ...Hantaviruses, a geographically diverse group of zoonotic pathogens, initiate cell infection through the concerted action of Gn and Gc viral surface glycoproteins. Here, we describe the high-resolution crystal structure of the antigenic ectodomain of Gn from Puumala hantavirus (PUUV), a causative agent of hemorrhagic fever with renal syndrome. Fitting of PUUV Gn into an electron cryomicroscopy reconstruction of intact Gn-Gc spike complexes from the closely related but non-pathogenic Tula hantavirus localized Gn tetramers to the membrane-distal surface of the virion. The accuracy of the fitting was corroborated by epitope mapping and genetic analysis of available PUUV sequences. Interestingly, Gn exhibits greater non-synonymous sequence diversity than the less accessible Gc, supporting a role of the host humoral immune response in exerting selective pressure on the virus surface. The fold of PUUV Gn is likely to be widely conserved across hantaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3364.map.gz emd_3364.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3364-v30.xml emd-3364-v30.xml emd-3364.xml emd-3364.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3364_fsc.xml emd_3364_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_3364.tif emd_3364.tif | 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3364 http://ftp.pdbj.org/pub/emdb/structures/EMD-3364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3364 | HTTPS FTP |
-Validation report
| Summary document |  emd_3364_validation.pdf.gz emd_3364_validation.pdf.gz | 271 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3364_full_validation.pdf.gz emd_3364_full_validation.pdf.gz | 270.2 KB | Display | |
| Data in XML |  emd_3364_validation.xml.gz emd_3364_validation.xml.gz | 8.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3364 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3364 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3364 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3364 | HTTPS FTP |
-Related structure data
| Related structure data |  5fynMC  5fxuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3364.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3364.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-tomogram average of Tula virus surface | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Glycoprotein spike of Tula hantavirus
| Entire | Name: Glycoprotein spike of Tula hantavirus |
|---|---|
| Components |
|
-Supramolecule #1000: Glycoprotein spike of Tula hantavirus
| Supramolecule | Name: Glycoprotein spike of Tula hantavirus / type: sample / ID: 1000 / Oligomeric state: Lattice of GnGc tetramers / Number unique components: 1 |
|---|
-Supramolecule #1: Tula virus
| Supramolecule | Name: Tula virus / type: virus / ID: 1 / Name.synonym: Tula hantavirus / NCBI-ID: 37133 / Sci species name: Tula virus / Sci species strain: Moravia / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: Tula hantavirus |
|---|---|
| Host (natural) | Organism:  Microtus (meadow voles) / synonym: VERTEBRATES Microtus (meadow voles) / synonym: VERTEBRATES |
| Host system | Organism: Chlorocebus / Recombinant cell: Vero E6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 25 mM Tris, 75 mM NaCl |
|---|---|
| Grid | Details: Grids (Cflat CF-2/1-2C-T) were glow-discharged for 15 s. 10-nm gold particles were added. |
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber humidity: 80 % / Chamber temperature: 120 K / Instrument: GATAN CRYOPLUNGE 3 / Method: Blot for 3 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80 K / Max: 120 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 160,000 times magnification. |
| Specialist optics | Energy filter - Name: GIF QUANTUM LS / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | Super-resolution counting mode |
| Date | Aug 14, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 30 / Average electron dose: 60 e/Å2 Details: Each image is a tilt series of 19 movies, acquired at 5 degree intervals. Each movie consists of 8 frames. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.8 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 160000 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -45 ° / Tilt series - Axis1 - Max angle: 45 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name: Chimera, Segger |
| Details | Density was segmented in 6 segments and 1000 evenly rotated fits were considered for each segment. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation |
| Output model |  PDB-5fyn: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)