[English] 日本語
 Yorodumi
Yorodumi- EMDB-23949: The insulin receptor ectodomain in complex with a venom hybrid in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23949 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The insulin receptor ectodomain in complex with a venom hybrid insulin analog - "head" region | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of female gonad development / positive regulation of meiotic cell cycle / insulin-like growth factor II binding / positive regulation of developmental growth / male sex determination / exocrine pancreas development / insulin receptor complex / insulin-like growth factor I binding / insulin receptor activity / positive regulation of protein-containing complex disassembly ...regulation of female gonad development / positive regulation of meiotic cell cycle / insulin-like growth factor II binding / positive regulation of developmental growth / male sex determination / exocrine pancreas development / insulin receptor complex / insulin-like growth factor I binding / insulin receptor activity / positive regulation of protein-containing complex disassembly / cargo receptor activity / dendritic spine maintenance / insulin binding / negative regulation of NAD(P)H oxidase activity / negative regulation of glycogen catabolic process / PTB domain binding / adrenal gland development / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / activation of protein kinase activity / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / Insulin processing / neuronal cell body membrane / regulation of protein secretion / positive regulation of peptide hormone secretion / positive regulation of respiratory burst / positive regulation of receptor internalization / negative regulation of acute inflammatory response / Regulation of gene expression in beta cells / alpha-beta T cell activation / amyloid-beta clearance / regulation of amino acid metabolic process / regulation of embryonic development / negative regulation of respiratory burst involved in inflammatory response / insulin receptor substrate binding / negative regulation of protein secretion / positive regulation of dendritic spine maintenance / transport across blood-brain barrier / positive regulation of glycogen biosynthetic process / Synthesis, secretion, and deacylation of Ghrelin / epidermis development / regulation of protein localization to plasma membrane / fatty acid homeostasis / negative regulation of lipid catabolic process / negative regulation of gluconeogenesis / Signal attenuation / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / COPI-mediated anterograde transport / phosphatidylinositol 3-kinase binding / positive regulation of lipid biosynthetic process / heart morphogenesis / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / positive regulation of insulin receptor signaling pathway / nitric oxide-cGMP-mediated signaling / negative regulation of reactive oxygen species biosynthetic process / positive regulation of protein autophosphorylation / transport vesicle / Insulin receptor recycling / insulin-like growth factor receptor binding / dendrite membrane / neuron projection maintenance / endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of brown fat cell differentiation / positive regulation of protein metabolic process / NPAS4 regulates expression of target genes / activation of protein kinase B activity / positive regulation of glycolytic process / positive regulation of mitotic nuclear division / Insulin receptor signalling cascade / receptor-mediated endocytosis / positive regulation of nitric-oxide synthase activity / learning / positive regulation of cytokine production / positive regulation of long-term synaptic potentiation / acute-phase response / endosome lumen / Regulation of insulin secretion / positive regulation of D-glucose import / positive regulation of protein secretion / negative regulation of proteolysis / positive regulation of cell differentiation / regulation of transmembrane transporter activity / insulin receptor binding / positive regulation of MAP kinase activity / wound healing / receptor protein-tyrosine kinase / caveola / regulation of synaptic plasticity / negative regulation of protein catabolic process / cellular response to growth factor stimulus / hormone activity / receptor internalization / memory / positive regulation of neuron projection development / peptidyl-tyrosine phosphorylation / cellular response to insulin stimulus / cognition / positive regulation of protein localization to nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
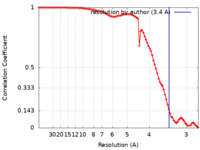
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Blakely AD / Xiong X / Kim JH / Menting J / Schafer IB / Schubert HL / Agrawal R / Gutmann T / Delaine C / Zhang Y ...Blakely AD / Xiong X / Kim JH / Menting J / Schafer IB / Schubert HL / Agrawal R / Gutmann T / Delaine C / Zhang Y / Artik GO / Merriman A / Eckert D / Lawrence MC / Coskun U / Fisher SJ / Forbes BE / Safavi-Hemami H / Hill CP / Chou DHC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2022 Journal: Nat Chem Biol / Year: 2022Title: Symmetric and asymmetric receptor conformation continuum induced by a new insulin. Authors: Xiaochun Xiong / Alan Blakely / Jin Hwan Kim / John G Menting / Ingmar B Schäfer / Heidi L Schubert / Rahul Agrawal / Theresia Gutmann / Carlie Delaine / Yi Wolf Zhang / Gizem Olay Artik / ...Authors: Xiaochun Xiong / Alan Blakely / Jin Hwan Kim / John G Menting / Ingmar B Schäfer / Heidi L Schubert / Rahul Agrawal / Theresia Gutmann / Carlie Delaine / Yi Wolf Zhang / Gizem Olay Artik / Allanah Merriman / Debbie Eckert / Michael C Lawrence / Ünal Coskun / Simon J Fisher / Briony E Forbes / Helena Safavi-Hemami / Christopher P Hill / Danny Hung-Chieh Chou /     Abstract: Cone snail venoms contain a wide variety of bioactive peptides, including insulin-like molecules with distinct structural features, binding modes and biochemical properties. Here, we report an active ...Cone snail venoms contain a wide variety of bioactive peptides, including insulin-like molecules with distinct structural features, binding modes and biochemical properties. Here, we report an active humanized cone snail venom insulin with an elongated A chain and a truncated B chain, and use cryo-electron microscopy (cryo-EM) and protein engineering to elucidate its interactions with the human insulin receptor (IR) ectodomain. We reveal how an extended A chain can compensate for deletion of B-chain residues, which are essential for activity of human insulin but also compromise therapeutic utility by delaying dissolution from the site of subcutaneous injection. This finding suggests approaches to developing improved therapeutic insulins. Curiously, the receptor displays a continuum of conformations from the symmetric state to a highly asymmetric low-abundance structure that displays coordination of a single humanized venom insulin using elements from both of the previously characterized site 1 and site 2 interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23949.map.gz emd_23949.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23949-v30.xml emd-23949-v30.xml emd-23949.xml emd-23949.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23949_fsc.xml emd_23949_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23949.png emd_23949.png | 106.7 KB | ||
| Masks |  emd_23949_msk_1.map emd_23949_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_23949_half_map_1.map.gz emd_23949_half_map_1.map.gz emd_23949_half_map_2.map.gz emd_23949_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23949 http://ftp.pdbj.org/pub/emdb/structures/EMD-23949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23949 | HTTPS FTP |
-Validation report
| Summary document |  emd_23949_validation.pdf.gz emd_23949_validation.pdf.gz | 703.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23949_full_validation.pdf.gz emd_23949_full_validation.pdf.gz | 703.3 KB | Display | |
| Data in XML |  emd_23949_validation.xml.gz emd_23949_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_23949_validation.cif.gz emd_23949_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23949 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23949 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23949 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23949 | HTTPS FTP |
-Related structure data
| Related structure data |  7mqoMC  7mqrC  7mqsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10736 (Title: Cryo-EM of the human insulin receptor ectodomain in complex with an insulin analog with truncated B chain and enlongated A chain EMPIAR-10736 (Title: Cryo-EM of the human insulin receptor ectodomain in complex with an insulin analog with truncated B chain and enlongated A chainData size: 11.7 TB Data #1: Unaligned multi-frame micrographs (40 e-/A2 dose) [micrographs - multiframe] Data #2: unaligned micrographs (60 e-/A2 dose) [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23949.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23949.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.365 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23949_msk_1.map emd_23949_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_23949_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_23949_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Insulin receptor ectodomain in complex with insulin analog Vh-Ins...
| Entire | Name: Insulin receptor ectodomain in complex with insulin analog Vh-Ins-HSLQ - focused refinement. |
|---|---|
| Components |
|
-Supramolecule #1: Insulin receptor ectodomain in complex with insulin analog Vh-Ins...
| Supramolecule | Name: Insulin receptor ectodomain in complex with insulin analog Vh-Ins-HSLQ - focused refinement. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 209 KDa |
-Macromolecule #1: Insulin A chain
| Macromolecule | Name: Insulin A chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.736106 KDa |
| Sequence | String: GIVEQCCTSI CSLYQLENYC HSLQ |
-Macromolecule #2: Insulin B chain
| Macromolecule | Name: Insulin B chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.537951 KDa |
| Sequence | String: FVNQHLCGSE LVEALYLVCL ER |
-Macromolecule #3: Isoform Short of Insulin receptor
| Macromolecule | Name: Isoform Short of Insulin receptor / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 104.761875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: HLYPGEVCPG MDIRNNLTRL HELENCSVIE GHLQILLMFK TRPEDFRDLS FPKLIMITDY LLLFRVYGLE SLKDLFPNLT VIRGSRLFF NYALVIFEMV HLKELGLYNL MNITRGSVRI EKNNELCYLA TIDWSRILDS VEDNYIVLNK DDNEECGDIC P GTAKGKTN ...String: HLYPGEVCPG MDIRNNLTRL HELENCSVIE GHLQILLMFK TRPEDFRDLS FPKLIMITDY LLLFRVYGLE SLKDLFPNLT VIRGSRLFF NYALVIFEMV HLKELGLYNL MNITRGSVRI EKNNELCYLA TIDWSRILDS VEDNYIVLNK DDNEECGDIC P GTAKGKTN CPATVINGQF VERCWTHSHC QKVCPTICKS HGCTAEGLCC HSECLGNCSQ PDDPTKCVAC RNFYLDGRCV ET CPPPYYH FQDWRCVNFS FCQDLHHKCK NSRRQGCHQY VIHNNKCIPE CPSGYTMNSS NLLCTPCLGP CPKVCHLLEG EKT IDSVTS AQELRGCTVI NGSLIINIRG GNNLAAELEA NLGLIEEISG YLKIRRSYAL VSLSFFRKLR LIRGETLEIG NYSF YALDN QNLRQLWDWS KHNLTITQGK LFFHYNPKLC LSEIHKMEEV SGTKGRQERN DIALKTNGDQ ASCENELLKF SYIRT SFDK ILLRWEPYWP PDFRDLLGFM LFYKEAPYQN VTEFDGQDAC GSNSWTVVDI DPPLRSNDPK SQNHPGWLMR GLKPWT QYA IFVKTLVTFS DERRTYGAKS DIIYVQTDAT NPSVPLDPIS VSNSSSQIIL KWKPPSDPNG NITHYLVFWE RQAEDSE LF ELDYCLKGLK LPSRTWSPPF ESEDSQKHNQ SEYEDSAGEC CSCPKTDSQI LKELEESSFR KTFEDYLHNV VFVPRPSR K RRSLGDVGNV TVAVPTVAAF PNTSSTSVPT SPEEHRPFEK VVNKESLVIS GLRHFTGYRI ELQACNQDTP EERCSVAAY VSARTMPEAK ADDIVGPVTH EIFENNVVHL MWQEPKEPNG LIVLYEVSYR RYGDEELHLC VSRKHFALER GCRLRGLSPG NYSVRIRAT SLAGNGSWTE PTYFYVTDYL DVPSNIAK |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Component: (Name: HEPES, Tris) Details: Equal parts HBS( 50 mM HEPES pH 7.5, 150 mM NaCl ) and TBS (25 mM Tris pH 8.5, 150 mM NaCl) |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)