+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22367 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human DPP9-CARD8 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CARD8 / DPP9 / inflammasome / Val-boroPro (VbP) / talabostat / innate immunity / IMMUNE SYSTEM / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationCARD8 inflammasome complex assembly / CARD8 inflammasome complex / NACHT domain binding / Formation of apoptosome / self proteolysis / CARD domain binding / NLRP3 inflammasome complex / negative regulation of NLRP3 inflammasome complex assembly / dipeptidyl-peptidase IV / negative regulation of lipopolysaccharide-mediated signaling pathway ...CARD8 inflammasome complex assembly / CARD8 inflammasome complex / NACHT domain binding / Formation of apoptosome / self proteolysis / CARD domain binding / NLRP3 inflammasome complex / negative regulation of NLRP3 inflammasome complex assembly / dipeptidyl-peptidase IV / negative regulation of lipopolysaccharide-mediated signaling pathway / cysteine-type endopeptidase activator activity / Regulation of the apoptosome activity / dipeptidyl-peptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / negative regulation of programmed cell death / negative regulation of interleukin-1 beta production / pattern recognition receptor activity / cell leading edge / pyroptotic inflammatory response / aminopeptidase activity / negative regulation of tumor necrosis factor-mediated signaling pathway / antiviral innate immune response / activation of innate immune response / intrinsic apoptotic signaling pathway / serine-type peptidase activity / negative regulation of canonical NF-kappaB signal transduction / positive regulation of interleukin-1 beta production / molecular condensate scaffold activity / peptidase activity / regulation of apoptotic process / defense response to virus / microtubule / inflammatory response / protein homodimerization activity / protein-containing complex / proteolysis / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
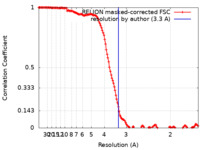
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Sharif H / Hollingsworth LR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2021 Journal: Immunity / Year: 2021Title: Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. Authors: Humayun Sharif / L Robert Hollingsworth / Andrew R Griswold / Jeffrey C Hsiao / Qinghui Wang / Daniel A Bachovchin / Hao Wu /  Abstract: CARD8 detects intracellular danger signals and forms a caspase-1 activating inflammasome. Like the related inflammasome sensor NLRP1, CARD8 autoprocesses into noncovalently associated N-terminal (NT) ...CARD8 detects intracellular danger signals and forms a caspase-1 activating inflammasome. Like the related inflammasome sensor NLRP1, CARD8 autoprocesses into noncovalently associated N-terminal (NT) and C-terminal (CT) fragments and binds the cellular dipeptidyl peptidases DPP8 and 9 (DPP8/9). Certain danger-associated signals, including the DPP8/9 inhibitor Val-boroPro (VbP) and HIV protease, induce proteasome-mediated NT degradation and thereby liberate the inflammasome-forming CT. Here, we report cryoelectron microscopy (cryo-EM) structures of CARD8 bound to DPP9, revealing a repressive ternary complex consisting of DPP9, full-length CARD8, and CARD8-CT. Unlike NLRP1-CT, CARD8-CT does not interact with the DPP8/9 active site and is not directly displaced by VbP. However, larger DPP8/9 active-site probes can directly weaken this complex in vitro, and VbP itself nevertheless appears to disrupt this complex, perhaps indirectly, in cells. Thus, DPP8/9 inhibitors can activate the CARD8 inflammasome by promoting CARD8 NT degradation and by weakening ternary complex stability. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22367.map.gz emd_22367.map.gz | 15.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22367-v30.xml emd-22367-v30.xml emd-22367.xml emd-22367.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22367_fsc.xml emd_22367_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22367.png emd_22367.png | 192.8 KB | ||
| Filedesc metadata |  emd-22367.cif.gz emd-22367.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22367 http://ftp.pdbj.org/pub/emdb/structures/EMD-22367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22367 | HTTPS FTP |
-Related structure data
| Related structure data |  7jkqMC  7jn7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10597 (Title: Human DPP9-CARD8 complex / Data size: 1.6 TB EMPIAR-10597 (Title: Human DPP9-CARD8 complex / Data size: 1.6 TBData #1: Unaligned multi frame micographs of CARD8-DPP9-noTILT [micrographs - multiframe] Data #2: Unaligned multi frame micographs of CARD8-DPP9-apo-TILT [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22367.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22367.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DPP9-CARD8 complex
| Entire | Name: DPP9-CARD8 complex |
|---|---|
| Components |
|
-Supramolecule #1: DPP9-CARD8 complex
| Supramolecule | Name: DPP9-CARD8 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Dipeptidyl peptidase 9
| Macromolecule | Name: Dipeptidyl peptidase 9 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: dipeptidyl-peptidase IV |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.38432 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATTGTPTAD RGDAAATDDP AARFQVQKHS WDGLRSIIHG SRKYSGLIVN KAPHDFQFVQ KTDESGPHSH RLYYLGMPYG SRENSLLYS EIPKKVRKEA LLLLSWKQML DHFQATPHHG VYSREEELLR ERKRLGVFGI TSYDFHSESG LFLFQASNSL F HCRDGGKN ...String: MATTGTPTAD RGDAAATDDP AARFQVQKHS WDGLRSIIHG SRKYSGLIVN KAPHDFQFVQ KTDESGPHSH RLYYLGMPYG SRENSLLYS EIPKKVRKEA LLLLSWKQML DHFQATPHHG VYSREEELLR ERKRLGVFGI TSYDFHSESG LFLFQASNSL F HCRDGGKN GFMVSPMKPL EIKTQCSGPR MDPKICPADP AFFSFINNSD LWVANIETGE ERRLTFCHQG LSNVLDDPKS AG VATFVIQ EEFDRFTGYW WCPTASWEGS EGLKTLRILY EEVDESEVEV IHVPSPALEE RKTDSYRYPR TGSKNPKIAL KLA EFQTDS QGKIVSTQEK ELVQPFSSLF PKVEYIARAG WTRDGKYAWA MFLDRPQQWL QLVLLPPALF IPSTENEEQR LASA RAVPR NVQPYVVYEE VTNVWINVHD IFYPFPQSEG EDELCFLRAN ECKTGFCHLY KVTAVLKSQG YDWSEPFSPG EDEFK CPIK EEIALTSGEW EVLARHGSKI WVNEETKLVY FQGTKDTPLE HHLYVVSYEA AGEIVRLTTP GFSHSCSMSQ NFDMFV SHY SSVSTPPCVH VYKLSGPDDD PLHKQPRFWA SMMEAASCPP DYVPPEIFHF HTRSDVRLYG MIYKPHALQP GKKHPTV LF VYGGPQVQLV NNSFKGIKYL RLNTLASLGY AVVVIDGRGS CQRGLRFEGA LKNQMGQVEI EDQVEGLQFV AEKYGFID L SRVAIHGWSY GGFLSLMGLI HKPQVFKVAI AGAPVTVWMA YDTGYTERYM DVPENNQHGY EAGSVALHVE KLPNEPNRL LILHGFLDEN VHFFHTNFLV SQLIRAGKPY QLQIYPNERH SIRCPESGEH YEVTLLHFLQ EYL UniProtKB: Dipeptidyl peptidase 9 |
-Macromolecule #2: Caspase recruitment domain-containing protein 8
| Macromolecule | Name: Caspase recruitment domain-containing protein 8 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.716875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEKKECPEKS SSSEEELPRR DSGSSRNIDA SKLIRLQGSR KLLVDNSIRE LQYTKTGIFF QAEACVTNDT VYRELPCVSE TLCDISHFF QEDDETEAEP LLFRAVPECQ LSGGDIPSVS EEQESSEGQD SGDICSEENQ IVSSYASKVC FEIEEDYKNR Q FLGPEGNV ...String: MEKKECPEKS SSSEEELPRR DSGSSRNIDA SKLIRLQGSR KLLVDNSIRE LQYTKTGIFF QAEACVTNDT VYRELPCVSE TLCDISHFF QEDDETEAEP LLFRAVPECQ LSGGDIPSVS EEQESSEGQD SGDICSEENQ IVSSYASKVC FEIEEDYKNR Q FLGPEGNV DVELIDKSTN RYSVWFPTAG WYLWSATGLG FLVRDEVTVT IAFGSWSQHL ALDLQHHEQW LVGGPLFDVT AE PEEAVAE IHLPHFISLQ AGEVDVSWFL VAHFKNEGMV LEHPARVEPF YAVLESPSFS LMGILLRIAS GTRLSIPITS NTL IYYHPH PEDIKFHLYL VPSDALLTKA IDDEEDRFHG VRLQTSPPME PLNFGSSYIV SNSANLKVMP KELKLSYRSP GEIQ HFSKF YAGQMKEPIQ LEITEKRHGT LVWDTEVKPV DLQLVAASAP PPFSGAAFVK ENHRQLQARM GDLKGVLDDL QDNEV LTEN EKELVEQEKT RQSKNEALLS MVEKKGDLAL DVLFRSISER DPYLVSYLRQ QNL UniProtKB: Caspase recruitment domain-containing protein 8 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25mM HEPES, pH 7.5, 150 mM NaCl, 1 mM TCEP |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #0 - Number grids imaged: 4 / #0 - Number real images: 3306 / #0 - Average exposure time: 2.22 sec. / #0 - Average electron dose: 58.5 e/Å2 / #0 - Details: stage tilt 0 degrees / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #1 - Number grids imaged: 4 / #1 - Number real images: 2488 / #1 - Average exposure time: 2.25 sec. / #1 - Average electron dose: 64.99 e/Å2 / #1 - Details: stage tilt 37 degrees |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Calibrated magnification: 10500 / Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: -0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)