[English] 日本語
 Yorodumi
Yorodumi- EMDB-9589: Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein an... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9589 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein and ACE2 complex, ACE2-free conformation with one RBD in up conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV / spike / glycoprotein / Class I fusion protein / membrane fusion / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / SARS-CoV-1 activates/modulates innate immune responses / symbiont-mediated-mediated suppression of host tetherin activity / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell ...Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / SARS-CoV-1 activates/modulates innate immune responses / symbiont-mediated-mediated suppression of host tetherin activity / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  SARS coronavirus / SARS coronavirus /  | |||||||||
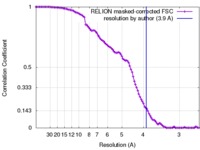
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Gui M / Song W | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2018 Journal: PLoS Pathog / Year: 2018Title: Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. Authors: Wenfei Song / Miao Gui / Xinquan Wang / Ye Xiang /  Abstract: The trimeric SARS coronavirus (SARS-CoV) surface spike (S) glycoprotein consisting of three S1-S2 heterodimers binds the cellular receptor angiotensin-converting enzyme 2 (ACE2) and mediates fusion ...The trimeric SARS coronavirus (SARS-CoV) surface spike (S) glycoprotein consisting of three S1-S2 heterodimers binds the cellular receptor angiotensin-converting enzyme 2 (ACE2) and mediates fusion of the viral and cellular membranes through a pre- to postfusion conformation transition. Here, we report the structure of the SARS-CoV S glycoprotein in complex with its host cell receptor ACE2 revealed by cryo-electron microscopy (cryo-EM). The complex structure shows that only one receptor-binding domain of the trimeric S glycoprotein binds ACE2 and adopts a protruding "up" conformation. In addition, we studied the structures of the SARS-CoV S glycoprotein and its complexes with ACE2 in different in vitro conditions, which may mimic different conformational states of the S glycoprotein during virus entry. Disassociation of the S1-ACE2 complex from some of the prefusion spikes was observed and characterized. We also characterized the rosette-like structures of the clustered SARS-CoV S2 trimers in the postfusion state observed on electron micrographs. Structural comparisons suggested that the SARS-CoV S glycoprotein retains a prefusion architecture after trypsin cleavage into the S1 and S2 subunits and acidic pH treatment. However, binding to the receptor opens up the receptor-binding domain of S1, which could promote the release of the S1-ACE2 complex and S1 monomers from the prefusion spike and trigger the pre- to postfusion conformational transition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9589.map.gz emd_9589.map.gz | 76.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9589-v30.xml emd-9589-v30.xml emd-9589.xml emd-9589.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9589_fsc.xml emd_9589_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9589.png emd_9589.png | 140.9 KB | ||
| Masks |  emd_9589_msk_1.map emd_9589_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9589.cif.gz emd-9589.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9589 http://ftp.pdbj.org/pub/emdb/structures/EMD-9589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9589 | HTTPS FTP |
-Validation report
| Summary document |  emd_9589_validation.pdf.gz emd_9589_validation.pdf.gz | 591.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9589_full_validation.pdf.gz emd_9589_full_validation.pdf.gz | 590.8 KB | Display | |
| Data in XML |  emd_9589_validation.xml.gz emd_9589_validation.xml.gz | 11.7 KB | Display | |
| Data in CIF |  emd_9589_validation.cif.gz emd_9589_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9589 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9589 | HTTPS FTP |
-Related structure data
| Related structure data |  6acdMC  9583C  9584C  9585C  9586C  9587C  9588C  9591C  9593C  9594C  9595C  9596C  9597C  9598C  6accC  6acgC  6acjC  6ackC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9589.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9589.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
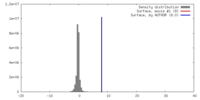
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9589_msk_1.map emd_9589_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
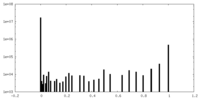
| Density Histograms |
- Sample components
Sample components
-Entire : Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein
| Entire | Name: Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein
| Supramolecule | Name: Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  SARS coronavirus SARS coronavirus |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133.763422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFIFLLFLTL TSGSDLDRCT TFDDVQAPNY TQHTSSMRGV YYPDEIFRSD TLYLTQDLFL PFYSNVTGFH TINHTFGNPV IPFKDGIYF AATEKSNVVR GWVFGSTMNN KSQSVIIINN STNVVIRACN FELCDNPFFA VSKPMGTQTH TMIFDNAFNC T FEYISDAF ...String: MFIFLLFLTL TSGSDLDRCT TFDDVQAPNY TQHTSSMRGV YYPDEIFRSD TLYLTQDLFL PFYSNVTGFH TINHTFGNPV IPFKDGIYF AATEKSNVVR GWVFGSTMNN KSQSVIIINN STNVVIRACN FELCDNPFFA VSKPMGTQTH TMIFDNAFNC T FEYISDAF SLDVSEKSGN FKHLREFVFK NKDGFLYVYK GYQPIDVVRD LPSGFNTLKP IFKLPLGINI TNFRAILTAF SP AQDIWGT SAAAYFVGYL KPTTFMLKYD ENGTITDAVD CSQNPLAELK CSVKSFEIDK GIYQTSNFRV VPSGDVVRFP NIT NLCPFG EVFNATKFPS VYAWERKKIS NCVADYSVLY NSTFFSTFKC YGVSATKLND LCFSNVYADS FVVKGDDVRQ IAPG QTGVI ADYNYKLPDD FMGCVLAWNT RNIDATSTGN YNYKYRYLRH GKLRPFERDI SNVPFSPDGK PCTPPALNCY WPLND YGFY TTTGIGYQPY RVVVLSFELL NAPATVCGPK LSTDLIKNQC VNFNFNGLTG TGVLTPSSKR FQPFQQFGRD VSDFTD SVR DPKTSEILDI SPCSFGGVSV ITPGTNASSE VAVLYQDVNC TDVSTAIHAD QLTPAWRIYS TGNNVFQTQA GCLIGAE HV DTSYECDIPI GAGICASYHT VSLLRSTSQK SIVAYTMSLG ADSSIAYSNN TIAIPTNFSI SITTEVMPVS MAKTSVDC N MYICGDSTEC ANLLLQYGSF CTQLNRALSG IAAEQDRNTR EVFAQVKQMY KTPTLKYFGG FNFSQILPDP LKPTKRSFI EDLLFNKVTL ADAGFMKQYG ECLGDINARD LICAQKFNGL TVLPPLLTDD MIAAYTAALV SGTATAGWTF GAGAALQIPF AMQMAYRFN GIGVTQNVLY ENQKQIANQF NKAISQIQES LTTTSTALGK LQDVVNQNAQ ALNTLVKQLS SNFGAISSVL N DILSRLDK VEAEVQIDRL ITGRLQSLQT YVTQQLIRAA EIRASANLAA TKMSECVLGQ SKRVDFCGKG YHLMSFPQAA PH GVVFLHV TYVPSQERNF TTAPAICHEG KAYFPREGVF VFNGTSWFIT QRNFFSPQII TTDNTFVSGN CDVVIGIINN TVY DPLQPE LDSFKEELDK YFKNHTSPDV DLGDISGINA SVVNIQKEID RLNEVAKNLN ESLIDLQELG KYEQYIKWPW SHPQ FEK UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 5.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 8.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)