+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4219 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Teneurin3 monomer | |||||||||
 Map data Map data | Teneurin3 monomer map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Neuronal cell adhesion / bacterial toxin-like / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of homophilic cell adhesion / synaptic membrane adhesion / homophilic cell-cell adhesion / presynaptic active zone membrane / positive regulation of neuron projection development / cell differentiation / protein heterodimerization activity / axon / glutamatergic synapse / signal transduction ...regulation of homophilic cell adhesion / synaptic membrane adhesion / homophilic cell-cell adhesion / presynaptic active zone membrane / positive regulation of neuron projection development / cell differentiation / protein heterodimerization activity / axon / glutamatergic synapse / signal transduction / protein homodimerization activity / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
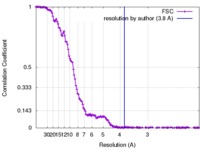
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Janssen BJC / Meijer DHM | |||||||||
| Funding support |  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction. Authors: Verity A Jackson / Dimphna H Meijer / Maria Carrasquero / Laura S van Bezouwen / Edward D Lowe / Colin Kleanthous / Bert J C Janssen / Elena Seiradake /   Abstract: Teneurins are ancient cell-cell adhesion receptors that are vital for brain development and synapse organisation. They originated in early metazoan evolution through a horizontal gene transfer event ...Teneurins are ancient cell-cell adhesion receptors that are vital for brain development and synapse organisation. They originated in early metazoan evolution through a horizontal gene transfer event when a bacterial YD-repeat toxin fused to a eukaryotic receptor. We present X-ray crystallography and cryo-EM structures of two Teneurins, revealing a ~200 kDa extracellular super-fold in which eight sub-domains form an intricate structure centred on a spiralling YD-repeat shell. An alternatively spliced loop, which is implicated in homophilic Teneurin interaction and specificity, is exposed and thus poised for interaction. The N-terminal side of the shell is 'plugged' via a fibronectin-plug domain combination, which defines a new class of YD proteins. Unexpectedly, we find that these proteins are widespread amongst modern bacteria, suggesting early metazoan receptor evolution from a distinct class of proteins, which today includes both bacterial proteins and eukaryotic Teneurins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4219.map.gz emd_4219.map.gz | 195.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4219-v30.xml emd-4219-v30.xml emd-4219.xml emd-4219.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4219_fsc.xml emd_4219_fsc.xml | 16 KB | Display |  FSC data file FSC data file |
| Images |  emd_4219.png emd_4219.png | 132 KB | ||
| Filedesc metadata |  emd-4219.cif.gz emd-4219.cif.gz | 7.1 KB | ||
| Others |  emd_4219_half_map_1.map.gz emd_4219_half_map_1.map.gz emd_4219_half_map_2.map.gz emd_4219_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4219 http://ftp.pdbj.org/pub/emdb/structures/EMD-4219 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4219 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4219 | HTTPS FTP |
-Related structure data
| Related structure data |  6fayMC  6fb3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4219.map.gz / Format: CCP4 / Size: 206 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4219.map.gz / Format: CCP4 / Size: 206 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Teneurin3 monomer map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half-map 1 for Teneurin3 monomer
| File | emd_4219_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 for Teneurin3 monomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
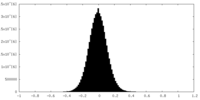
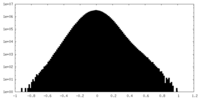
| Density Histograms |
-Half map: Half-map 2 for Teneurin3 monomer
| File | emd_4219_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 for Teneurin3 monomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
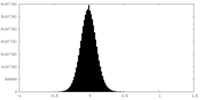
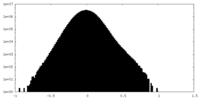
| Density Histograms |
- Sample components
Sample components
-Entire : Teneurin3 monomer
| Entire | Name: Teneurin3 monomer |
|---|---|
| Components |
|
-Supramolecule #1: Teneurin3 monomer
| Supramolecule | Name: Teneurin3 monomer / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 220 KDa |
-Macromolecule #1: Odz3 protein
| Macromolecule | Name: Odz3 protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 211.429734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSFYDRISFL IGSDSTHVLP GESPFNKSLA SVIRGQVLTA DGTPLIGVNV SFLHYSEYGY TITRQDGMFD LVANGGASLT LVFERSPFL TQYHTVWIPW NVFYVMDTLV MKKEENDIPS CDLSGFVRPS PIIVSSPLST FFRSSPEDSP IIPETQVLHE E TTIPGTDL ...String: GSFYDRISFL IGSDSTHVLP GESPFNKSLA SVIRGQVLTA DGTPLIGVNV SFLHYSEYGY TITRQDGMFD LVANGGASLT LVFERSPFL TQYHTVWIPW NVFYVMDTLV MKKEENDIPS CDLSGFVRPS PIIVSSPLST FFRSSPEDSP IIPETQVLHE E TTIPGTDL KLSYLSSRAA GYKSVLKITM TQAVIPFNLM KVHLMVAVVG RLFQKWFPAS PNLAYTFIWD KTDAYNQKVY GL SEAVVSV GYEYESCLDL TLWEKRTAVL QGYELDASNM GGWTLDKHHV LDVQNGILYK GNGENQFISQ QPPVVSSIMG NGR RRSISC PSCNGQADGN KLLAPVALAC GIDGSLYVGD FNYVRRIFPS GNVTSVLELS SNPAHRYYLA TDPVTGDLYV SDTN TRRIY RPKSLTGAKD LTKNAEVVAG TGEQCLPFDE ARCGDGGKAV EATLMSPKGM AIDKNGLIYF VDGTMIRKVD QNGII STLL GSNDLTSARP LTCDTSMHIS QVRLEWPTDL AINPMDNSIY VLDNNVVLQI TENRQVRIAA GRPMHCQVPG VEYPVG KHA VQTTLESATA IAVSYSGVLY ITETDEKKIN RIRQVTTDGE ISLVAGIPSE CDCKNDANCD CYQSGDGYAK DAKLNAP SS LAASPDGTLY IADLGNIRIR AVSKNKPLLN SMNFYEVASP TDQELYIFDI NGTHQYTVSL VTGDYLYNFS YSNDNDVT A VTDSNGNTLR IRRDPNRMPV RVVSPDNQVI WLTIGTNGCL KSMTAQGLEL VLFTYHGNSG LLATKSDETG WTTFFDYDS EGRLTNVTFP TGVVTNLHGD MDKAITVDIE SSSREEDVSI TSNLSSIDSF YTMVQDQLRN SYQIGYDGSL RIFYASGLDS HYQTEPHVL AGTANPTVAK RNMTLPGENG QNLVEWRFRK EQAQGKVNVF GRKLRVNGRN LLSVDFDRTT KTEKIYDDHR K FLLRIAYD TSGHPTLWLP SSKLMAVNVT YSSTGQIASI QRGTTSEKVD YDSQGRIVSR VFADGKTWSY TYLEKSMVLL LH SQRQYIF EYDMWDRLSA ITMPSVARHT MQTIRSIGYY RNIYNPPESN ASIITDYNEE GLLLQTAFLG TSRRVLFKYR RQT RLSEIL YDSTRVSFTY DETAGVLKTV NLQSDGFICT IRYRQIGPLI DRQIFRFSED GMVNARFDYS YDNSFRVTSM QGVI NETPL PIDLYQFDDI SGKVEQFGKF GVIYYDINQI ISTAVMTYTK HFDAHGRIKE IQYEIFRSLM YWITIQYDNM GRVTK REIK IGPFANTTKY AYEYDVDGQL QTVYLNEKIM WRYNYDLNGN LHLLNPSSSA RLTPLRYDLR DRITRLGDVQ YRLDED GFL RQRGTEIFEY SSKGLLTRVY SKGSGWTVIY RYDGLGRRVS SKTSLGQHLQ FFYADLTYPT RITHVYNHSS SEITSLY YD LQGHLFAMEI SSGDEFYIAS DNTGTPLAVF SSNGLMLKQI QYTAYGEIYF DSNVDFQLVI GFHGGLYDPL TKLIHFGE R DYDILAGRWT TPDIEIWKRI GKDPAPFNLY MFRNNNPASK IHDVKDYITD VNSWLVTFGF HLHNAIPGFP VPKFDLTEP SYELVKSQQW EDVPPIFGVQ QQVARQAKAF LSLGKMAEVQ VSRRKAGAEQ SWLWFATVKS LIGKGVMLAV SQGRVQTNVL NIANEDCIK VAAVLNNAFY LENLHFTIEG KDTHYFIKTT TPESDLGTLR LTSGRKALEN GINVTVSQST TVVNGRTRRF A DVEMQFGA LALHVRYGMT LDEEKARILE QARQRALARA WAREQQRVRD GEEGARLWTE GEKRQLLSAG KVQGYDGYYV LS VEQYPEL ADSANNIQFL RQSEIGKRAS AAAHHHHHH UniProtKB: Teneurin-3 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 9 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model Details: We have used PDB 6FB3 (same publication) as a template model using Modeller v9.16 followed by manual rebuilding in Coot and real-space refinement using Phenix. |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 126 |
| Output model |  PDB-6fay: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)