+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3063 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Slo2.2 Na+-activated K+ channel | |||||||||
 Map data Map data | Focus refined reconstruction of chicken Slo2.2 gating ring | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / potassium channel | |||||||||
| Function / homology |  Function and homology information Function and homology informationintracellular sodium-activated potassium channel activity / outward rectifier potassium channel activity / potassium ion transmembrane transport / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
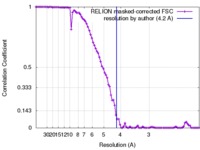
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Hite RK / Yuan P / Li Z / Hsuing Y / Walz T / MacKinnon R | |||||||||
 Citation Citation |  Journal: Nature / Year: 2015 Journal: Nature / Year: 2015Title: Cryo-electron microscopy structure of the Slo2.2 Na(+)-activated K(+) channel. Authors: Richard K Hite / Peng Yuan / Zongli Li / Yichun Hsuing / Thomas Walz / Roderick MacKinnon /  Abstract: Na(+)-activated K(+) channels are members of the Slo family of large conductance K(+) channels that are widely expressed in the brain, where their opening regulates neuronal excitability. These ...Na(+)-activated K(+) channels are members of the Slo family of large conductance K(+) channels that are widely expressed in the brain, where their opening regulates neuronal excitability. These channels fulfil a number of biological roles and have intriguing biophysical properties, including conductance levels that are ten times those of most other K(+) channels and gating sensitivity to intracellular Na(+). Here we present the structure of a complete Na(+)-activated K(+) channel, chicken Slo2.2, in the Na(+)-free state, determined by cryo-electron microscopy at a nominal resolution of 4.5 ångströms. The channel is composed of a large cytoplasmic gating ring, in which resides the Na(+)-binding site and a transmembrane domain that closely resembles voltage-gated K(+) channels. In the structure, the cytoplasmic domain adopts a closed conformation and the ion conduction pore is also closed. The structure reveals features that can explain the unusually high conductance of Slo channels and how contraction of the cytoplasmic gating ring closes the pore. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3063.map.gz emd_3063.map.gz | 56.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3063-v30.xml emd-3063-v30.xml emd-3063.xml emd-3063.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3063_fsc.xml emd_3063_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_3063.png emd_3063.png | 146.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3063 http://ftp.pdbj.org/pub/emdb/structures/EMD-3063 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3063 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3063 | HTTPS FTP |
-Validation report
| Summary document |  emd_3063_validation.pdf.gz emd_3063_validation.pdf.gz | 275.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3063_full_validation.pdf.gz emd_3063_full_validation.pdf.gz | 274.5 KB | Display | |
| Data in XML |  emd_3063_validation.xml.gz emd_3063_validation.xml.gz | 10.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3063 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3063 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3063 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3063 | HTTPS FTP |
-Related structure data
| Related structure data |  5a6fMC  3062C  3064C  5a6eC  5a6gC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3063.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3063.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focus refined reconstruction of chicken Slo2.2 gating ring | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : chicken Slo2.2
| Entire | Name: chicken Slo2.2 |
|---|---|
| Components |
|
-Supramolecule #1000: chicken Slo2.2
| Supramolecule | Name: chicken Slo2.2 / type: sample / ID: 1000 / Oligomeric state: tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 550 KDa |
-Macromolecule #1: Slo2.2
| Macromolecule | Name: Slo2.2 / type: protein_or_peptide / ID: 1 / Name.synonym: Slack, KCNT1 / Number of copies: 4 / Oligomeric state: tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 550 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Potassium channel subfamily T member 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 20 mM HEPES pH 7.4, 300 mM KCl, 1.5 mM dodecyl maltoside, 0.05 mg/ml POPE:POPG (3:1) |
| Grid | Details: 300 mesh Cu Quantifoil R1.2 / 1.3 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 84 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK IV / Method: 4 second blot prior to plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: FEI Image corrector |
| Specialist optics | Energy filter - Name: Gatan / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 30.0 eV |
| Date | Jul 7, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 2243 / Average electron dose: 40 e/Å2 Details: 25 sub-frames were recorded for each image in super-resolution counting mode. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)