[English] 日本語
 Yorodumi
Yorodumi- EMDB-24847: Locally refined protomer structure of native-form oocyte/egg Alph... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24847 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Locally refined protomer structure of native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer | |||||||||
 Map data Map data | Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Xenopus egg extract / Protease inhibitor / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species | ||||||||||
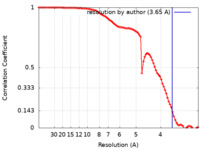
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Arimura Y / Funabiki H | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2022 Journal: J Mol Biol / Year: 2022Title: Structural Mechanics of the Alpha-2-Macroglobulin Transformation. Authors: Yasuhiro Arimura / Hironori Funabiki /  Abstract: Alpha-2-Macroglobulin (A2M) is the critical pan-protease inhibitor of the innate immune system. When proteases cleave the A2M bait region, global structural transformation of the A2M tetramer is ...Alpha-2-Macroglobulin (A2M) is the critical pan-protease inhibitor of the innate immune system. When proteases cleave the A2M bait region, global structural transformation of the A2M tetramer is triggered to entrap the protease. The structural basis behind the cleavage-induced transformation and the protease entrapment remains unclear. Here, we report cryo-EM structures of native- and intermediate-forms of the Xenopus laevis egg A2M homolog (A2Moo or ovomacroglobulin) tetramer at 3.7-4.1 Å and 6.4 Å resolution, respectively. In the native A2Moo tetramer, two pairs of dimers arrange into a cross-like configuration with four 60 Å-wide bait-exposing grooves. Each bait in the native form threads into an aperture formed by three macroglobulin domains (MG2, MG3, MG6). The bait is released from the narrowed aperture in the induced protomer of the intermediate form. We propose that the intact bait region works as a "latch-lock" to block futile A2M transformation until its protease-mediated cleavage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24847.map.gz emd_24847.map.gz | 56.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24847-v30.xml emd-24847-v30.xml emd-24847.xml emd-24847.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24847_fsc.xml emd_24847_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_24847.png emd_24847.png | 130 KB | ||
| Filedesc metadata |  emd-24847.cif.gz emd-24847.cif.gz | 6.3 KB | ||
| Others |  emd_24847_additional_1.map.gz emd_24847_additional_1.map.gz emd_24847_half_map_1.map.gz emd_24847_half_map_1.map.gz emd_24847_half_map_2.map.gz emd_24847_half_map_2.map.gz | 59.5 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24847 http://ftp.pdbj.org/pub/emdb/structures/EMD-24847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24847 | HTTPS FTP |
-Related structure data
| Related structure data |  7s62MC  7s63C  7s64C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24847.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24847.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.47 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer
| File | emd_24847_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer | ||||||||||||
| Projections & Slices |
| ||||||||||||
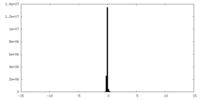
| Density Histograms |
-Half map: Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer
| File | emd_24847_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer | ||||||||||||
| Projections & Slices |
| ||||||||||||
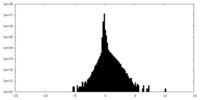
| Density Histograms |
-Half map: Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer
| File | emd_24847_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Native-form oocyte/egg Alpha-2-Macroglobulin (A2Moo) tetramer | ||||||||||||
| Projections & Slices |
| ||||||||||||
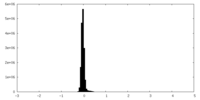
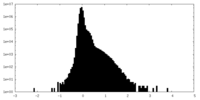
| Density Histograms |
- Sample components
Sample components
-Entire : nucleosome isolated from metaphase chromosome formed in Xenopus e...
| Entire | Name: nucleosome isolated from metaphase chromosome formed in Xenopus egg extract (oligo fraction) |
|---|---|
| Components |
|
-Supramolecule #1: nucleosome isolated from metaphase chromosome formed in Xenopus e...
| Supramolecule | Name: nucleosome isolated from metaphase chromosome formed in Xenopus egg extract (oligo fraction) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: Alpha 2-macroglobulin
| Macromolecule | Name: Alpha 2-macroglobulin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 159.480812 KDa |
| Sequence | String: MSPNRFLLCV CILGLIAGGA AKVQYALTIP ALLKSGETQR ACVNLIGYHQ PLALSVVLEH QRVNISIFSE KVQPPHYFKC NKFMVPTVI TNAPDFVTLS VSGGGEDIKD RKAVVIAPLN TICLIQMDKP VYKPGCKVRF RLISLNTMLL PISEKYTAVY L EDPSGSRI ...String: MSPNRFLLCV CILGLIAGGA AKVQYALTIP ALLKSGETQR ACVNLIGYHQ PLALSVVLEH QRVNISIFSE KVQPPHYFKC NKFMVPTVI TNAPDFVTLS VSGGGEDIKD RKAVVIAPLN TICLIQMDKP VYKPGCKVRF RLISLNTMLL PISEKYTAVY L EDPSGSRI AQWQNQESVG GVVQLEFPLI SDAAPGSYTI TAEGESCESA RQGFTVDEYI LPRFSVIVDP PNTISILDDI LT LNVSAIY TYGQPVPGSV TIKCCREASS YYGRKGNCFK GNRGICTNIT GELGPDGAFY GVVSLLPFQM GQSGFQMSLG VAL TVTEEG TGIQVTHQFF IMITSQLATL IFDYDALKEF YKRGIPYLVK VILTDANDNP MANEQVEVEL AGKTIGAVLT DKEG RAEYA IDTSSFVQEN FTVVVSYENP HQCYYTEWEG PDFPTAQHFV MRFYSETGSF LDIQGSSVEL NCGQVHNISV RYILS LDGM GEGATTATFY YLAMSRAKIV QHGQRDVHLN QSKSGLFNIG LNVTSDLAPG AELIVYCILD LELIADTISL DIEKCF QNQ VSLSFSDDLG PTASNVSLNL SAAPGSLCGV KVIDSSLLLI NPYESLSASG VYYSIPYLSL FGYNYGGFNL EEPEPPC ED PNTVIFCKGR YYLPVSSSTE GDTYQNLRRV GLVLGTSSKI RKPVVCGMEA KFSVPRKSSG ESDFGSSLSN GHVETLRK N FSETFLWRLV SVDSEGQNTI TETVPDTITK WQGSMFCVSE KEGFGITKYS ANFTSFLPFF VELSLPYSLT REEILVMKA FVSNYLEECI KIIVTLQPSA DFEVIPQDVK QDQCICSGGR SSYSWNIIAS SLGRISFIVS AETTHIGASC DGPSDQSQST RKDTVIQTI LVQPEGIRKE ETSSNLVCVE DSNVEMPINL TLPENIVQGS ASAFVTFVGD VLGLPLSNLQ NLLQMPYGCG E QNLARMAP IPYVLEYLNN TNQLTDELLQ TAVQFLNEGY YRQLRYKLPS GAYDAFWSSP SDGSSWLSAY TFKTFEKAKK YI YVDGKIQ QQTLLYLQTS QKLDNGCFKA EGNLFMRQCG QERDLCFTAY LAIALLESNY SSGMTLLDDA LGCLEAAMSS AST LYFKSY TVYVFTLVQN WEIRNTLLNE LKSKVVSERG TLHWEREDKL GQEGIPLYYP NYSPAEVEIT AYMLLSIAKG SDPT HDDLT YMAQISVWLI QQQNSYGGFR STQDTVVALQ ALAFYAQLLF KSNAHHNVFL RSEYGDVGQL NLSEHNRLVV QRLQL PEVS GNYSISINGT GCCLVQSTIR YNIPVPKENS AFYVAADSVS KNCLNGVAYT ITITVSVSYR GLRNETNMVI IDIQML SGY QADYPSLRQL ENSQQVSKTE EQDNHVFLYL NAVPLKTIQL SFKVLIGSRV LNVKSASVYV YDYYETGENG FASYSQP C UniProtKB: Uncharacterized protein LOC431886 isoform X1 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 17 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: SUPER-RESOLUTION / #0 - Average electron dose: 33.11 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Detector mode: SUPER-RESOLUTION / #1 - Average electron dose: 38.34 e/Å2 / #2 - Image recording ID: 3 / #2 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #2 - Detector mode: SUPER-RESOLUTION / #2 - Average electron dose: 35.27 e/Å2 / #3 - Image recording ID: 4 / #3 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #3 - Detector mode: SUPER-RESOLUTION / #3 - Average electron dose: 34.2 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)