[English] 日本語
 Yorodumi
Yorodumi- EMDB-23909: Structural basis for CSPG4 as a receptor for TcdB and a therapeut... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23909 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection | |||||||||
 Map data Map data | Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CSPG4 / TcdB / Clostridioides difficile infection / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationCS-GAG biosynthesis / Defective CHST3 causes SEDCJD / Defective CHST14 causes EDS, musculocontractural type / Defective CHSY1 causes TPBS / DS-GAG biosynthesis / CS/DS degradation / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / substrate-dependent cell migration / Defective B3GAT3 causes JDSSDHD ...CS-GAG biosynthesis / Defective CHST3 causes SEDCJD / Defective CHST14 causes EDS, musculocontractural type / Defective CHSY1 causes TPBS / DS-GAG biosynthesis / CS/DS degradation / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / substrate-dependent cell migration / Defective B3GAT3 causes JDSSDHD / symbiont-mediated perturbation of host actin cytoskeleton via filamentous actin depolymerization / Glycosaminoglycan-protein linkage region biosynthesis / glial cell migration / tissue remodeling / ruffle assembly / glucosyltransferase activity / Differentiation of Keratinocytes in Interfollicular Epidermis in Mammalian Skin / positive regulation of peptidyl-tyrosine phosphorylation / Transferases; Glycosyltransferases; Hexosyltransferases / host cell cytosol / platelet-derived growth factor receptor signaling pathway / lamellipodium membrane / coreceptor activity / ruffle / cysteine-type peptidase activity / lysosomal lumen / host cell endosome membrane / Golgi lumen / : / toxin activity / angiogenesis / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / positive regulation of MAPK cascade / intracellular signal transduction / apical plasma membrane / focal adhesion / lipid binding / protein kinase binding / host cell plasma membrane / cell surface / proteolysis / extracellular exosome / extracellular region / nucleoplasm / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Clostridioides difficile (bacteria) / Clostridioides difficile (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
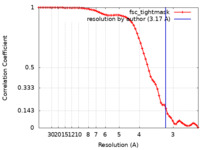
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Chen P / Jin R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection. Authors: Peng Chen / Ji Zeng / Zheng Liu / Hatim Thaker / Siyu Wang / Songhai Tian / Jie Zhang / Liang Tao / Craig B Gutierrez / Li Xing / Ralf Gerhard / Lan Huang / Min Dong / Rongsheng Jin /    Abstract: C. difficile is a major cause of antibiotic-associated gastrointestinal infections. Two C. difficile exotoxins (TcdA and TcdB) are major virulence factors associated with these infections, and ...C. difficile is a major cause of antibiotic-associated gastrointestinal infections. Two C. difficile exotoxins (TcdA and TcdB) are major virulence factors associated with these infections, and chondroitin sulfate proteoglycan 4 (CSPG4) is a potential receptor for TcdB, but its pathophysiological relevance and the molecular details that govern recognition remain unknown. Here, we determine the cryo-EM structure of a TcdB-CSPG4 complex, revealing a unique binding site spatially composed of multiple discontinuous regions across TcdB. Mutations that selectively disrupt CSPG4 binding reduce TcdB toxicity in mice, while CSPG4-knockout mice show reduced damage to colonic tissues during C. difficile infections. We further show that bezlotoxumab, the only FDA approved anti-TcdB antibody, blocks CSPG4 binding via an allosteric mechanism, but it displays low neutralizing potency on many TcdB variants from epidemic hypervirulent strains due to sequence variations in its epitopes. In contrast, a CSPG4-mimicking decoy neutralizes major TcdB variants, suggesting a strategy to develop broad-spectrum therapeutics against TcdB. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23909.map.gz emd_23909.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23909-v30.xml emd-23909-v30.xml emd-23909.xml emd-23909.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23909_fsc.xml emd_23909_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_23909.png emd_23909.png | 106.2 KB | ||
| Filedesc metadata |  emd-23909.cif.gz emd-23909.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23909 http://ftp.pdbj.org/pub/emdb/structures/EMD-23909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23909 | HTTPS FTP |
-Validation report
| Summary document |  emd_23909_validation.pdf.gz emd_23909_validation.pdf.gz | 602.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23909_full_validation.pdf.gz emd_23909_full_validation.pdf.gz | 602.5 KB | Display | |
| Data in XML |  emd_23909_validation.xml.gz emd_23909_validation.xml.gz | 9.8 KB | Display | |
| Data in CIF |  emd_23909_validation.cif.gz emd_23909_validation.cif.gz | 12.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23909 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23909 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23909 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23909 | HTTPS FTP |
-Related structure data
| Related structure data |  7ml7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23909.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23909.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EM map of TcdB in complex with CSPG4
| Entire | Name: EM map of TcdB in complex with CSPG4 |
|---|---|
| Components |
|
-Supramolecule #1: EM map of TcdB in complex with CSPG4
| Supramolecule | Name: EM map of TcdB in complex with CSPG4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: TcdB
| Supramolecule | Name: TcdB / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
-Supramolecule #3: CSPG4
| Supramolecule | Name: CSPG4 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Toxin B
| Macromolecule | Name: Toxin B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
| Molecular weight | Theoretical: 228.752531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAWSHPQFE KGGGSGGGSG GSAWSHPQFE KLEVLFQGPM SLVNRKQLEK MANVRFRTQE DEYVAILDAL EEYHNMSENT VVEKYLKLK DINSLTDIYI DTYKKSGRNK ALKKFKEYLV TEVLELKNNN LTPVEKNLHF VAIGGQINDT AINYINQWKD V NSDYNVNV ...String: MSAWSHPQFE KGGGSGGGSG GSAWSHPQFE KLEVLFQGPM SLVNRKQLEK MANVRFRTQE DEYVAILDAL EEYHNMSENT VVEKYLKLK DINSLTDIYI DTYKKSGRNK ALKKFKEYLV TEVLELKNNN LTPVEKNLHF VAIGGQINDT AINYINQWKD V NSDYNVNV FYDSNAFLIN TLKKTVVESA INDTLESFRE NLNDPRFDYN KFFRKRMEII YDKQKNFINY YKAQREENPE LI IDDIVKT YLSNEYSKEI DELNTYIEES LNKITQNSGN DVRNFEEFKN GESFNLYEQE LVERWNLAAA SDILRISALK EIG GMYLNV NMLPGIQPDL FESIEKPSSV TVDFWEMTKL EAIMKYKEYI PEYTSEHFDM LDEEVQSSFE SVLASKSDKS EIFS SLGDM EASPLEVKIA FNSKGIINQG LISVKDSYCS NLIVKQIENR YKILNNSLNP AISEDNDFNT TTNTFIDSIM AEANA DNGR FMMELGKYLR VGFFPDVKTT INLSGPEAYA AAYQDLLMFK EGSMNIHLIE ADLRNFEISK TNISQSTEQE MASLWS FDD ARAKAQFEEY KRNYFEGSAG EDDNLDFSQN IVVDKEYLLE KISSLARSSE RGYIHYIVQL QGDKISYEAA CNLFAKT PY DSVLFQKNIE DSEIAYYYNP GDGEIQEIDK YKIPSIISDR PKIKLTFIGH GKDEFNTDIF AGFDVDSLST EIEAAIDL A KEDISPKSIE INLLGCNMFS YSINVEETYP GKLLLKVKDK ISELMPSISQ DSIIVSANQY EVRINSEGRR ELLDHSGEW INKEESIIKD ISSKEYISFN PKENKITVKS KNLPELSTLL QEIRNNSNSS DIELEEKVML TECEINVISN IDTQIVEERI EEAKNLTSD SINYIKDEFK LIESISDALC DLKQQNELED SHFISFEDIS ETDEGFSIRF INKETGESIF VETEKTIFSE Y ANHITEEI SKIKGTIFDT VNGKLVKKVN LDTTHEVNTL NAAFFIQSLI EYNSSKESLS NLSVAMKVQV YAQLFSTGLN TI TDAAKVV ELVSTALDET IDLLPTLSEG LPIIATIIDG VSLGAAIKEL SETSDPLLRQ EIEAKIGIMA VNLTTATTAI ITS SLGIAS GFSILLVPLA GISAGIPSLV NNELVLRDKA TKVVDYFKHV SLVETEGVFT LLDDKIMMPQ DDLVISEIDF NNNS IVLGK CEIWRMEGGS GHTVTDDIDH FFSAPSITYR EPHLSIYDVL EVQKEELDLS KDLMVLPNAP NRVFAWETGW TPGLR SLEN DGTKLLDRIR DNYEGEFYWR YFAFIADALI TTLKPRYEDT NIRINLDSNT RSFIVPIITT EYIREKLSYS FYGSGG TYA LSLSQYNMGI NIELSESDVW IIDVDNVVRD VTIESDKIKK GDLIEGILST LSIEENKIIL NSHEINFSGE VNGSNGF VS LTFSILEGIN AIIEVDLLSK SYKLLISGEL KILMLNSNHI QQKIDYIGFN SELQKNIPYS FVDSEGKENG FINGSTKE G LFVSELPDVV LISKVYMDDS KPSFGYYSNN LKDVKVITKD NVNILTGYYL KDDIKISLSL TLQDEKTIKL NSVHLDESG VAEILKFMNR KGNTNTSDSL MSFLESMNIK SIFVNFLQSN IKFILDANFI ISGTTSIGQF EFICDENDNI QPYFIKFNTL ETNYTLYVG NRQNMIVEPN YDLDDSGDIS STVINFSQKY LYGIDSCVNK VVISPNIYTD EINITPVYET NNTYPEVIVL D ANYINEKI NVNINDLSIR YVWSNDGNDF ILMSTSEENK VSQVKIRFVN VFKDKTLANK LSFNFSDKQD VPVSEIILSF TP SYYEDGL IGYDLGLVSL YNEKFYINNF GMMVSGLIYI NDSLYYFKPP VNNLITGFVT VGDDKYYFNP INGGAASIGE TII DDKNYY FNQSGVLQTG VFSTEDGFKY FAPANTLDEN LEGEAIDFTG KLIIDENIYY FDDNYRGAVE WKELDGEMHY FSPE TGKAF KLEHHHHHH UniProtKB: Toxin B |
-Macromolecule #2: Chondroitin sulfate proteoglycan 4
| Macromolecule | Name: Chondroitin sulfate proteoglycan 4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.298305 KDa |
| Recombinant expression | Organism: Mammalian expression vector BsrGI-MCS-pcDNA3.1 (others) |
| Sequence | String: HHHHHHHHHS GGGSGGGIEG RPSGSASFFG ENHLEVPVAT ALTDIDLQLQ FSTSQPEALL LLAAGPADHL LLQLYSGRLQ VRLVLGQEE LRLQTPAETL LSDSIPHTVV LTVVEGWATL SVDGFLNASS AVPGAPLEVP YGLFVGGTGT LGLPYLRGTS R PLRGCLHA ...String: HHHHHHHHHS GGGSGGGIEG RPSGSASFFG ENHLEVPVAT ALTDIDLQLQ FSTSQPEALL LLAAGPADHL LLQLYSGRLQ VRLVLGQEE LRLQTPAETL LSDSIPHTVV LTVVEGWATL SVDGFLNASS AVPGAPLEVP YGLFVGGTGT LGLPYLRGTS R PLRGCLHA ATLNGRSLLR PLTPDVHEGC AEEFSASDDV ALGFSGPHSL AAFPAWGTQD EGTLEFTLTT QSRQAPLAFQ AG GRRGDFI YVDIFEGHLR AVVEKGQGTV LLHNSVPVAD GQPHEVSVHI NAHRLEISVD QYPTHTSNRG VLSYLEPRGS LLL GGLDAE ASRHLQEHRL GLTPEATNAS LLGCMEDLSV NGQRRGLREA LLTRNMAAGC RLEEEEYEDD AYGHYEAFST LAPE AWPAM ELPEPCVPEP GLPPVFANFT QLLTISPLVV AEGGTAWLEW RHVQPTLDLM EAELRKSQVL FSVTRGARHG ELELD IPGA QARKMFTLLD VVNRKARFIH DGSEDTSDQL VLEVSVTARV PMPSCLRRGQ TYLLPIQVNP VNDPPHIIFP HGSLMV ILE HTQKPLGPEV FQAYDPDSAC EGLTFQVLGT SSGLPVERRD QPGEPATEFS CRELEAGSLV YVHRGGPAQD LTFRVSD GL QASPPATLKV VAIRPAIQIH RSTGLRLAQG SAMPILPANL SVETNAVGQD VSVLFRVTGA LQFGELQKQG AGGVEGAE W WATQAFHQRD VEQGRVRYLS TDPQHHAYDT VENLALEVQ UniProtKB: Chondroitin sulfate proteoglycan 4 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)