+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20270 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the K. lactis CBF3 core | |||||||||
 Map data Map data | K. lactis CBF3 core | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | yeast centromere-binding complex / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-binding transcription activator activity, RNA polymerase II-specific / ubiquitin-dependent protein catabolic process / protein ubiquitination / RNA polymerase II cis-regulatory region sequence-specific DNA binding / zinc ion binding / membrane / nucleus Similarity search - Function | |||||||||
| Biological species |  Kluyveromyces lactis (yeast) Kluyveromyces lactis (yeast) | |||||||||
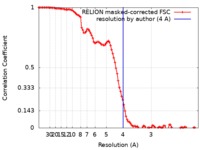
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Lee PD / Wei H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2019 Journal: J Mol Biol / Year: 2019Title: Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis. Authors: Phong D Lee / Hui Wei / Dongyan Tan / Stephen C Harrison /  Abstract: Kinetochores are the multiprotein complexes that link chromosomal centromeres to mitotic-spindle microtubules. Budding yeast centromeres comprise three sequential "centromere-determining elements", ...Kinetochores are the multiprotein complexes that link chromosomal centromeres to mitotic-spindle microtubules. Budding yeast centromeres comprise three sequential "centromere-determining elements", CDEI, II, and III. CDEI (8 bp) and CDEIII (∼25 bp) are conserved between Kluyveromyces lactis and Saccharomyces cerevisiae, but CDEII in the former is twice as long (160 bp) as CDEII in the latter (80 bp). The CBF3 complex recognizes CDEIII and is required for assembly of a centromeric nucleosome, which in turn recruits other kinetochore components. To understand differences in centromeric nucleosome assembly between K. lactis and S. cerevisiae, we determined the structure of a K. lactis CBF3 complex by electron cryomicroscopy at ∼4 Å resolution and compared it with published structures of S. cerevisiae CBF3. We show differences in the pose of Ndc10 and discuss potential models of the K. lactis centromeric nucleosome that account for the extended CDEII length. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20270.map.gz emd_20270.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20270-v30.xml emd-20270-v30.xml emd-20270.xml emd-20270.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20270_fsc.xml emd_20270_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_20270.png emd_20270.png | 63 KB | ||
| Filedesc metadata |  emd-20270.cif.gz emd-20270.cif.gz | 5.9 KB | ||
| Others |  emd_20270_half_map_1.map.gz emd_20270_half_map_1.map.gz emd_20270_half_map_2.map.gz emd_20270_half_map_2.map.gz | 71.2 MB 71.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20270 http://ftp.pdbj.org/pub/emdb/structures/EMD-20270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20270 | HTTPS FTP |
-Related structure data
| Related structure data |  6p7vMC  6p7wC  6p7xC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20270.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20270.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | K. lactis CBF3 core | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0605 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
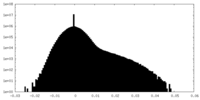
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: K. lactis CBF3 core, half map 1
| File | emd_20270_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | K. lactis CBF3 core, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
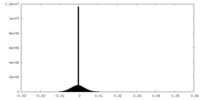
| Density Histograms |
-Half map: K. lactis CBF3 core, half map 2
| File | emd_20270_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | K. lactis CBF3 core, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
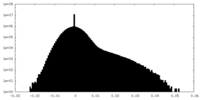
| Density Histograms |
- Sample components
Sample components
-Entire : K. lactis CBF3 core
| Entire | Name: K. lactis CBF3 core |
|---|---|
| Components |
|
-Supramolecule #1: K. lactis CBF3 core
| Supramolecule | Name: K. lactis CBF3 core / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Kluyveromyces lactis (yeast) Kluyveromyces lactis (yeast) |
-Macromolecule #1: Ctf13
| Macromolecule | Name: Ctf13 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Kluyveromyces lactis (yeast) Kluyveromyces lactis (yeast) |
| Molecular weight | Theoretical: 46.159945 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MFDTKLFLSL PIDIRYTVYF FLGDVVQNVR PPAKSDIFND ELIAYPNIRE FNQSLVDKYS KHIGVYDYIP NFIPNWCRDF DLLRHDIIL TDRLRVCLQY EEQWFSVQWI VVSGELEIGI FTTDEQFLQV SYTINEYCHL LSIAQQDLRL GINVSDINDV N ELCKEIQH ...String: MFDTKLFLSL PIDIRYTVYF FLGDVVQNVR PPAKSDIFND ELIAYPNIRE FNQSLVDKYS KHIGVYDYIP NFIPNWCRDF DLLRHDIIL TDRLRVCLQY EEQWFSVQWI VVSGELEIGI FTTDEQFLQV SYTINEYCHL LSIAQQDLRL GINVSDINDV N ELCKEIQH RWLFDTVSYI SFINCWDLDH ENVVSIIPCM ESFNNLHMLR IESKNMFNNL INTQGVRENP GKTIVYNVRQ NI FELELYT LRDLGYKSVV DLQKWEQLQC LSLSGCEFID LNNLILPQHC KMLILKEVKY IIWWDLSHLL KRIRPQWIIN GQV KKPTKK EEEEESEWYN LYLEVVQTYQ PLNFIELHNA KRVKGNLILP ARLVTESRIK ISNGTKVDSV LLI UniProtKB: KLLA0F13816p |
-Macromolecule #2: Skp1
| Macromolecule | Name: Skp1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Kluyveromyces lactis (yeast) Kluyveromyces lactis (yeast) |
| Molecular weight | Theoretical: 21.081141 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSKENQNVVL VSVEGERFVV DRKIAERSLL LKNYLQDLNS GDLHDDNDAD DDEDDEEDGD DEIVMPVPNV RSSVLQKVIE WAVHHKDSN FPDEDDDDSR KAAPVDPWDR EFLKVDQEML YEIILAANYL NIKPLLDAGC KVVAEMIRGR TPEEIRRTFN I VNDFTPEE EAAIRRENEW AEDR UniProtKB: E3 ubiquitin ligase complex SCF subunit |
-Macromolecule #3: Cep3
| Macromolecule | Name: Cep3 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Kluyveromyces lactis (yeast) Kluyveromyces lactis (yeast) |
| Molecular weight | Theoretical: 74.165312 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSKPKISLTK GKHPCTFCQA RKVKCDRSLP ACQNCIERNV TELCEYDDNG SRKRARLADD VNLYDKKLFN IWNQYERLWI HDTLGQCQQ GVYMGIAFPL DVSEYNNTKD FYGYECLFSK ESIFKILDHS LERLGWLYFG FFTDISELPY QMERYWNEYE S MNINLENE ...String: MSKPKISLTK GKHPCTFCQA RKVKCDRSLP ACQNCIERNV TELCEYDDNG SRKRARLADD VNLYDKKLFN IWNQYERLWI HDTLGQCQQ GVYMGIAFPL DVSEYNNTKD FYGYECLFSK ESIFKILDHS LERLGWLYFG FFTDISELPY QMERYWNEYE S MNINLENE EATTRQTTFK KSADQILWDL VLRSVIVMTI YYMPAKSILS LVDIDAIEKY PLDFSESNEG VDELKKKYEI FD YCLRHTL NKVLRTIFTL PPDVRTLQIF LILSNTNFLQ IYPSLGNNIL VHCIHLAKVL GIKDFKLKIN DSGSTRLQKL SMH NIWFRL STVDYMRSSP NKIIALHTDN SSALTRKTLF THCSIDSIDV YDVESNLEVL RWKITSLDRD LEVSEPSLKT LKAM KELLG LLDRKTSVSN DASFNTKFES FFLKLQCNFV MWKILRYEFM QYGVTNGLQK LCCPARRIIA LVANFLKEDY FEYTT HPFC VHILCVIAGF FSFYCIFHEA DEVRDLRNDA VGLLKLLFDP LRPVISCFFS NLSRLEELRH IWKSVEITDQ ANRLVH PVM YVLKTDIIKL KRNLEIISGS LKDANYQETF KDKLEIDINT PALSSDFLEV VREFNLSHPL DINGKMSRQN N UniProtKB: KLLA0D09977p |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)