[English] 日本語
 Yorodumi
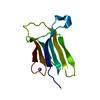
Yorodumi- PDB-1uqs: The Crystal Structure of Human CD1b with a Bound Bacterial Glycolipid -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1uqs | ||||||
|---|---|---|---|---|---|---|---|
| Title | The Crystal Structure of Human CD1b with a Bound Bacterial Glycolipid | ||||||
 Components Components |
| ||||||
 Keywords Keywords | GLYCOPROTEIN / LIPID / GMM / CD1B / MHC / ANTIGEN PRESENTATION | ||||||
| Function / homology |  Function and homology information Function and homology information: / : / : / : / : / exogenous lipid antigen binding / antigen processing and presentation, endogenous lipid antigen via MHC class Ib / lipopeptide binding / endogenous lipid antigen binding / regulation of membrane depolarization ...: / : / : / : / : / exogenous lipid antigen binding / antigen processing and presentation, endogenous lipid antigen via MHC class Ib / lipopeptide binding / endogenous lipid antigen binding / regulation of membrane depolarization / antigen processing and presentation, exogenous lipid antigen via MHC class Ib / retina homeostasis / positive regulation of protein binding / negative regulation of receptor binding / early endosome lumen / Nef mediated downregulation of MHC class I complex cell surface expression / DAP12 interactions / transferrin transport / cellular response to iron ion / Endosomal/Vacuolar pathway / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / peptide antigen assembly with MHC class II protein complex / cellular response to iron(III) ion / MHC class II protein complex / negative regulation of forebrain neuron differentiation / antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent / iron ion transport / ER to Golgi transport vesicle membrane / peptide antigen assembly with MHC class I protein complex / regulation of iron ion transport / regulation of erythrocyte differentiation / HFE-transferrin receptor complex / response to molecule of bacterial origin / MHC class I peptide loading complex / T cell mediated cytotoxicity / positive regulation of T cell cytokine production / antigen processing and presentation of endogenous peptide antigen via MHC class I / antigen processing and presentation of exogenous peptide antigen via MHC class II / positive regulation of immune response / MHC class I protein complex / positive regulation of T cell activation / peptide antigen binding / positive regulation of receptor-mediated endocytosis / negative regulation of neurogenesis / cellular response to nicotine / positive regulation of T cell mediated cytotoxicity / multicellular organismal-level iron ion homeostasis / specific granule lumen / phagocytic vesicle membrane / recycling endosome membrane / Interferon gamma signaling / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / negative regulation of epithelial cell proliferation / antimicrobial humoral immune response mediated by antimicrobial peptide / MHC class II protein complex binding / Modulation by Mtb of host immune system / late endosome membrane / sensory perception of smell / antibacterial humoral response / positive regulation of cellular senescence / tertiary granule lumen / DAP12 signaling / T cell differentiation in thymus / negative regulation of neuron projection development / cellular response to lipopolysaccharide / ER-Phagosome pathway / protein refolding / early endosome membrane / defense response to Gram-negative bacterium / protein homotetramerization / adaptive immune response / amyloid fibril formation / intracellular iron ion homeostasis / learning or memory / endosome membrane / defense response to Gram-positive bacterium / immune response / endoplasmic reticulum lumen / Amyloid fiber formation / Golgi membrane / lysosomal membrane / innate immune response / external side of plasma membrane / focal adhesion / intracellular membrane-bounded organelle / Neutrophil degranulation / SARS-CoV-2 activates/modulates innate and adaptive immune responses / structural molecule activity / cell surface / endoplasmic reticulum / Golgi apparatus / protein homodimerization activity / extracellular space / extracellular exosome / extracellular region / identical protein binding / membrane / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.1 Å MOLECULAR REPLACEMENT / Resolution: 3.1 Å | ||||||
 Authors Authors | Batuwangala, T. / Shepherd, D. / Gadola, S.D. / Gibson, K.J.C. / Zaccai, N.R. / Besra, G.S. / Cerundolo, V. / Jones, E.Y. | ||||||
 Citation Citation |  Journal: J Immunol. / Year: 2004 Journal: J Immunol. / Year: 2004Title: The crystal structure of human CD1b with a bound bacterial glycolipid. Authors: Batuwangala, T. / Shepherd, D. / Gadola, S.D. / Gibson, K.J. / Zaccai, N.R. / Fersht, A.R. / Besra, G.S. / Cerundolo, V. / Jones, E.Y. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN ... SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1uqs.cif.gz 1uqs.cif.gz | 90.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1uqs.ent.gz pdb1uqs.ent.gz | 68.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1uqs.json.gz 1uqs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1uqs_validation.pdf.gz 1uqs_validation.pdf.gz | 752.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1uqs_full_validation.pdf.gz 1uqs_full_validation.pdf.gz | 787.7 KB | Display | |
| Data in XML |  1uqs_validation.xml.gz 1uqs_validation.xml.gz | 23.1 KB | Display | |
| Data in CIF |  1uqs_validation.cif.gz 1uqs_validation.cif.gz | 30.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uq/1uqs https://data.pdbj.org/pub/pdb/validation_reports/uq/1uqs ftp://data.pdbj.org/pub/pdb/validation_reports/uq/1uqs ftp://data.pdbj.org/pub/pdb/validation_reports/uq/1uqs | HTTPS FTP |
-Related structure data
| Related structure data |  1gzqS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 33088.215 Da / Num. of mol.: 1 / Fragment: FRAGMENT: RESIDUES 18-295 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PET23D / Production host: HOMO SAPIENS (human) / Plasmid: PET23D / Production host:  |
|---|---|
| #2: Protein | Mass: 11879.356 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PET23D / Production host: HOMO SAPIENS (human) / Plasmid: PET23D / Production host:  |
| #3: Chemical | ChemComp-GMM / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.5 Å3/Da / Density % sol: 67 % / Description: WEISSENBERG METHOD | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 5.6 Details: 0.1M SODIUM CITRATE PH 5.6, 0.5M AMMONIUM SULFATE, 0.5M LITHIUM SULFATE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ / pH: 6.5 / Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 1.01 / Beamline: ID14-4 / Wavelength: 1.01 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Nov 15, 2002 / Details: MIRRORS |
| Radiation | Monochromator: SILICON CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.01 Å / Relative weight: 1 |
| Reflection | Resolution: 3.1→20 Å / Num. obs: 11697 / % possible obs: 99.9 % / Observed criterion σ(I): 1 / Redundancy: 12.8 % / Rmerge(I) obs: 0.077 / Net I/σ(I): 34.9 |
| Reflection shell | Resolution: 3.1→3.21 Å / Rmerge(I) obs: 0.604 / Mean I/σ(I) obs: 5.7 / % possible all: 100 |
| Reflection | *PLUS Highest resolution: 3.1 Å / Lowest resolution: 20 Å / Num. measured all: 150075 / Rmerge(I) obs: 0.077 |
| Reflection shell | *PLUS % possible obs: 100 % / Rmerge(I) obs: 0.604 / Mean I/σ(I) obs: 5.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1GZQ Resolution: 3.1→19.9 Å / Rfactor Rfree error: 0.01 / Data cutoff high absF: 1353771.84 / Isotropic thermal model: GROUP / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 28.6 Å2 / ksol: 0.194067 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 87 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.1→19.9 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.1→3.29 Å / Rfactor Rfree error: 0.041 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rfree: 0.291 / Rfactor Rwork: 0.233 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller





































































 PDBj
PDBj