[English] 日本語
 Yorodumi
Yorodumi- EMDB-11014: Cryo-EM map of human dihydrolipoamide succinyltransferase catalyt... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11014 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
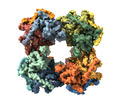
| Title | Cryo-EM map of human dihydrolipoamide succinyltransferase catalytic domain (DLST) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
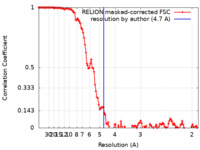
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Bailey HJ / Bezerra GA / Foster W / McCorvie TJ / Saez LD / Yue WW | |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2020 Journal: IUCrJ / Year: 2020Title: Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism. Authors: Gustavo A Bezerra / William R Foster / Henry J Bailey / Kevin G Hicks / Sven W Sauer / Bianca Dimitrov / Thomas J McCorvie / Jürgen G Okun / Jared Rutter / Stefan Kölker / Wyatt W Yue /    Abstract: DHTKD1 is a lesser-studied E1 enzyme among the family of 2-oxoacid de-hydrogenases. In complex with E2 (di-hydro-lipo-amide succinyltransferase, DLST) and E3 (dihydrolipo-amide de-hydrogenase, DLD) ...DHTKD1 is a lesser-studied E1 enzyme among the family of 2-oxoacid de-hydrogenases. In complex with E2 (di-hydro-lipo-amide succinyltransferase, DLST) and E3 (dihydrolipo-amide de-hydrogenase, DLD) components, DHTKD1 is involved in lysine and tryptophan catabolism by catalysing the oxidative de-carboxyl-ation of 2-oxoadipate (2OA) in mitochondria. Here, the 1.9 Å resolution crystal structure of human DHTKD1 is solved in complex with the thi-amine diphosphate co-factor. The structure reveals how the DHTKD1 active site is modelled upon the well characterized homologue 2-oxoglutarate (2OG) de-hydrogenase but engineered specifically to accommodate its preference for the longer substrate of 2OA over 2OG. A 4.7 Å resolution reconstruction of the human DLST catalytic core is also generated by single-particle electron microscopy, revealing a 24-mer cubic scaffold for assembling DHTKD1 and DLD protomers into a megacomplex. It is further demonstrated that missense DHTKD1 variants causing the inborn error of 2-amino-adipic and 2-oxoadipic aciduria impact on the complex formation, either directly by disrupting the interaction with DLST, or indirectly through destabilizing the DHTKD1 protein. This study provides the starting framework for developing DHTKD1 modulators to probe the intricate mitochondrial energy metabolism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11014.map.gz emd_11014.map.gz | 13.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11014-v30.xml emd-11014-v30.xml emd-11014.xml emd-11014.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11014_fsc.xml emd_11014_fsc.xml | 12.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_11014.png emd_11014.png | 185.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11014 http://ftp.pdbj.org/pub/emdb/structures/EMD-11014 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11014 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11014 | HTTPS FTP |
-Validation report
| Summary document |  emd_11014_validation.pdf.gz emd_11014_validation.pdf.gz | 250.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11014_full_validation.pdf.gz emd_11014_full_validation.pdf.gz | 249.4 KB | Display | |
| Data in XML |  emd_11014_validation.xml.gz emd_11014_validation.xml.gz | 12.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11014 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11014 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11014 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11014 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11014.map.gz / Format: CCP4 / Size: 155.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11014.map.gz / Format: CCP4 / Size: 155.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human dihydrolipoamide succinyltransferase (DLST)
| Entire | Name: human dihydrolipoamide succinyltransferase (DLST) |
|---|---|
| Components |
|
-Supramolecule #1: human dihydrolipoamide succinyltransferase (DLST)
| Supramolecule | Name: human dihydrolipoamide succinyltransferase (DLST) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.059 MDa |
-Macromolecule #1: human dihydrolipoamide succinyltransferase catalytic domain (DLST)
| Macromolecule | Name: human dihydrolipoamide succinyltransferase catalytic domain (DLST) type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHSS GVDLGTENLY FQSMDDLVTV KTPAFAESVT EGDVRWEKAV GDTVAEDEVV CEIETDKTSV QVPSPANGVI EALLVPDGGK VEGGTPLFTL RKTGAAPAKA KPAEAPAAAA PKAEPTAAAV PPPAAPIPTQ MPPVPSPSQP PSGKPVSAVK PTVAPPLAEP ...String: MGHHHHHHSS GVDLGTENLY FQSMDDLVTV KTPAFAESVT EGDVRWEKAV GDTVAEDEVV CEIETDKTSV QVPSPANGVI EALLVPDGGK VEGGTPLFTL RKTGAAPAKA KPAEAPAAAA PKAEPTAAAV PPPAAPIPTQ MPPVPSPSQP PSGKPVSAVK PTVAPPLAEP GAGKGLRSEH REKMNRMRQR IAQRLKEAQN TCAMLTTFNE IDMSNIQEMR ARHKEAFLKK HNLKLGFMSA FVKASAFALQ EQPVVNAVID DTTKEVVYRD YIDISVAVAT PRGLVVPVIR NVEAMNFADI ERTITELGEK ARKNELAIED MDGGTFTISN GGVFGSLFGT PIINPPQSAI LGMHGIFDRP VAIGGKVEVR PMMYVALTYD HRLIDGREAV TFLRKIKAAV EDPRVLLLDL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: OTHER / Number grids imaged: 1 / Number real images: 619 / Average exposure time: 1.0 sec. / Average electron dose: 32.52 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal magnification: 150000 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)