[English] 日本語
 Yorodumi
Yorodumi- EMDB-0108: Cryo-electron microscopic structure of the dihydrolipoamide succi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0108 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
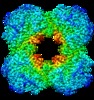
| Title | Cryo-electron microscopic structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate (2-oxoglutarate) dehydrogenase complex [residues 218-453] | |||||||||||||||||||||
 Map data Map data | Cryo-electron microscopic map of the human dihydrolipoamide succinyltransferase [residues 218-453] | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | alpha-ketoglutarate dehydrogenase complex / 2-oxoglutarate dehydrogenase complex / dihydrolipoamide succinyltransferase / E2 component / TRANSFERASE | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information2-oxoglutarate decarboxylation to succinyl-CoA / OGDH complex synthesizes succinyl-CoA from 2-OG / OADH complex synthesizes glutaryl-CoA from 2-OA / oxoadipate dehydrogenase complex / Glycine degradation / L-lysine catabolic process to acetyl-CoA via L-saccharopine / succinyl-CoA metabolic process / dihydrolipoyllysine-residue succinyltransferase / dihydrolipoyllysine-residue succinyltransferase activity / oxoglutarate dehydrogenase complex ...2-oxoglutarate decarboxylation to succinyl-CoA / OGDH complex synthesizes succinyl-CoA from 2-OG / OADH complex synthesizes glutaryl-CoA from 2-OA / oxoadipate dehydrogenase complex / Glycine degradation / L-lysine catabolic process to acetyl-CoA via L-saccharopine / succinyl-CoA metabolic process / dihydrolipoyllysine-residue succinyltransferase / dihydrolipoyllysine-residue succinyltransferase activity / oxoglutarate dehydrogenase complex / Protein lipoylation / 2-oxoglutarate metabolic process / acyltransferase activity / tricarboxylic acid cycle / generation of precursor metabolites and energy / mitochondrial matrix / mitochondrion / nucleoplasm / membrane / nucleus / cytosol Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||||||||
 Authors Authors | Nagy B / Zambo Z | |||||||||||||||||||||
| Funding support |  Hungary, Hungary,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Biochim Biophys Acta Gen Subj / Year: 2021 Journal: Biochim Biophys Acta Gen Subj / Year: 2021Title: Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: ...Title: Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: Implications for the overall hKGDHc structure. Authors: Balint Nagy / Martin Polak / Oliver Ozohanics / Zsofia Zambo / Eszter Szabo / Agnes Hubert / Frank Jordan / Jiří Novaček / Vera Adam-Vizi / Attila Ambrus /    Abstract: BACKGROUND: The human mitochondrial alpha-ketoglutarate dehydrogenase complex (hKGDHc) converts KG to succinyl-CoA and NADH. Malfunction of and reactive oxygen species generation by the hKGDHc as ...BACKGROUND: The human mitochondrial alpha-ketoglutarate dehydrogenase complex (hKGDHc) converts KG to succinyl-CoA and NADH. Malfunction of and reactive oxygen species generation by the hKGDHc as well as its E1-E2 subcomplex are implicated in neurodegenerative disorders, ischemia-reperfusion injury, E3-deficiency and cancers. METHODS: We performed cryo-EM, cross-linking mass spectrometry (CL-MS) and molecular modeling analyses to determine the structure of the E2 component of the hKGDHc (hE2k); hE2k transfers a succinyl ...METHODS: We performed cryo-EM, cross-linking mass spectrometry (CL-MS) and molecular modeling analyses to determine the structure of the E2 component of the hKGDHc (hE2k); hE2k transfers a succinyl group to CoA and forms the structural core of hKGDHc. We also assessed the overall structure of the hKGDHc by negative-stain EM and modeling. RESULTS: We report the 2.9 Å resolution cryo-EM structure of the hE2k component. The cryo-EM map comprises density for hE2k residues 151-386 - the entire (inner) core catalytic domain plus a few ...RESULTS: We report the 2.9 Å resolution cryo-EM structure of the hE2k component. The cryo-EM map comprises density for hE2k residues 151-386 - the entire (inner) core catalytic domain plus a few additional residues -, while residues 1-150 are not observed due to the inherent flexibility of the N-terminal region. The structure of the latter segment was also determined by CL-MS and homology modeling. Negative-stain EM on in vitro assembled hKGDHc and previous data were used to build a putative overall structural model of the hKGDHc. CONCLUSIONS: The E2 core of the hKGDHc is composed of 24 hE2k chains organized in octahedral (8 × 3 type) assembly. Each lipoyl domain is oriented towards the core domain of an adjacent chain in ...CONCLUSIONS: The E2 core of the hKGDHc is composed of 24 hE2k chains organized in octahedral (8 × 3 type) assembly. Each lipoyl domain is oriented towards the core domain of an adjacent chain in the hE2k homotrimer. hE1k and hE3 are most likely tethered at the edges and faces, respectively, of the cubic hE2k assembly. GENERAL SIGNIFICANCE: The revealed structural information will support the future pharmacologically targeting of the hKGDHc. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0108.map.gz emd_0108.map.gz | 78.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0108-v30.xml emd-0108-v30.xml emd-0108.xml emd-0108.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0108.png emd_0108.png | 264.6 KB | ||
| Filedesc metadata |  emd-0108.cif.gz emd-0108.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0108 http://ftp.pdbj.org/pub/emdb/structures/EMD-0108 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0108 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0108 | HTTPS FTP |
-Related structure data
| Related structure data |  6h05MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0108.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0108.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-electron microscopic map of the human dihydrolipoamide succinyltransferase [residues 218-453] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Affinity purified human dihydrolipoamide succinyltransferase
| Entire | Name: Affinity purified human dihydrolipoamide succinyltransferase |
|---|---|
| Components |
|
-Supramolecule #1: Affinity purified human dihydrolipoamide succinyltransferase
| Supramolecule | Name: Affinity purified human dihydrolipoamide succinyltransferase type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Protein in 100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 8.0. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Dihydrolipoyllysine-residue succinyltransferase component of 2-ox...
| Macromolecule | Name: Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial type: protein_or_peptide / ID: 1 Details: N-terminal Twin-Strep (affinity) tag with proteolytic cleavage site and linkers: MASWSHPQFEKGGGSGGGSGGSAWSHPQFEKLEVLFQGPG Density was found for 236 residues [218-453] Number of copies: 1 / Enantiomer: LEVO / EC number: dihydrolipoyllysine-residue succinyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.543004 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASWSHPQFE KGGGSGGGSG GSAWSHPQFE KLEVLFQGPG DDLVTVKTPA FAESVTEGDV RWEKAVGDTV AEDEVVCEIE TDKTSVQVP SPANGVIEAL LVPDGGKVEG GTPLFTLRKT GAAPAKAKPA EAPAAAAPKA EPTAAAVPPP AAPIPTQMPP V PSPSQPPS ...String: MASWSHPQFE KGGGSGGGSG GSAWSHPQFE KLEVLFQGPG DDLVTVKTPA FAESVTEGDV RWEKAVGDTV AEDEVVCEIE TDKTSVQVP SPANGVIEAL LVPDGGKVEG GTPLFTLRKT GAAPAKAKPA EAPAAAAPKA EPTAAAVPPP AAPIPTQMPP V PSPSQPPS GKPVSAVKPT VAPPLAEPGA GKGLRSEHRE KMNRMRQRIA QRLKEAQNTC AMLTTFNEID MSNIQEMRAR HK EAFLKKH NLKLGFMSAF VKASAFALQE QPVVNAVIDD TTKEVVYRDY IDISVAVATP RGLVVPVIRN VEAMNFADIE RTI TELGEK ARKNELAIED MDGGTFTISN GGVFGSLFGT PIINPPQSAI LGMHGIFDRP VAIGGKVEVR PMMYVALTYD HRLI DGREA VTFLRKIKAA VEDPRVLLLD L UniProtKB: Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | This sample was highly purified and monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 48.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | COOT was used for structure manipulations, while MOLPROBITY was applied for validation. |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-6h05: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















