[English] 日本語
 Yorodumi
Yorodumi- EMDB-10556: Cryo-EM map of human dihydrolipoamide succinyltransferase catalyt... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10556 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of human dihydrolipoamide succinyltransferase catalytic domain (DLST) | |||||||||
 Map data Map data | Human dihydrolipoamide succinyltransferase (E2) component of the 2-oxoglutarate dehydrogenase complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
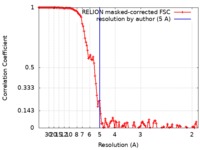
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | |||||||||
 Authors Authors | Bailey HJ / Bezerra GA / Foster W / Diaz Saez L / Yue WW | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM map of human dihydrolipoamide succinyltransferase (DLST) Authors: Bailey HJ / Bezerra GA / Foster W / Diaz Saez L / Yue WW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10556.map.gz emd_10556.map.gz | 16.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10556-v30.xml emd-10556-v30.xml emd-10556.xml emd-10556.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10556_fsc.xml emd_10556_fsc.xml | 12.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10556.png emd_10556.png | 136.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10556 http://ftp.pdbj.org/pub/emdb/structures/EMD-10556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10556 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10556.map.gz / Format: CCP4 / Size: 155.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10556.map.gz / Format: CCP4 / Size: 155.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human dihydrolipoamide succinyltransferase (E2) component of the 2-oxoglutarate dehydrogenase complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human dihydrolipoamide succinyltransferase (DLST)
| Entire | Name: human dihydrolipoamide succinyltransferase (DLST) |
|---|---|
| Components |
|
-Supramolecule #1: human dihydrolipoamide succinyltransferase (DLST)
| Supramolecule | Name: human dihydrolipoamide succinyltransferase (DLST) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.059 MDa |
-Macromolecule #1: human dihydrolipoamide succinyltransferase (DLST)
| Macromolecule | Name: human dihydrolipoamide succinyltransferase (DLST) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHSS GVDLGTENLY FQSMDDLVTV KTPAFAESVT EGDVRWEKAV GDTVAEDEVV CEIETDKTSV QVPSPANGVI EALLVPDGGK VEGGTPLFTL RKTGAAPAKA KPAEAPAAAA PKAEPTAAAV PPPAAPIPTQ MPPVPSPSQP PSGKPVSAVK PTVAPPLAEP ...String: MGHHHHHHSS GVDLGTENLY FQSMDDLVTV KTPAFAESVT EGDVRWEKAV GDTVAEDEVV CEIETDKTSV QVPSPANGVI EALLVPDGGK VEGGTPLFTL RKTGAAPAKA KPAEAPAAAA PKAEPTAAAV PPPAAPIPTQ MPPVPSPSQP PSGKPVSAVK PTVAPPLAEP GAGKGLRSEH REKMNRMRQR IAQRLKEAQN TCAMLTTFNE IDMSNIQEMR ARHKEAFLKK HNLKLGFMSA FVKASAFALQ EQPVVNAVID DTTKEVVYRD YIDISVAVAT PRGLVVPVIR NVEAMNFADI ERTITELGEK ARKNELAIED MDGGTFTISN GGVFGSLFGT PIINPPQSAI LGMHGIFDRP VAIGGKVEVR PMMYVALTYD HRLIDGREAV TFLRKIKAAV EDPRVLLLDL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: OTHER / Number grids imaged: 1 / Number real images: 619 / Average exposure time: 1.0 sec. / Average electron dose: 32.52 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal magnification: 150000 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)