-Search query
-Search result
Showing 1 - 50 of 71 items for (author: zhou & jm)

EMDB-35827: 
Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate

EMDB-37652: 
Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate

EMDB-37656: 
Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary)

EMDB-37657: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary)

EMDB-37762: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex)

EMDB-43753: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain

EMDB-41374: 
Antibody N3-1 bound to RBDs in the up and down conformations
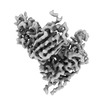
EMDB-41382: 
Antibody N3-1 bound to RBD in the up conformation
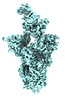
EMDB-41399: 
Antibody N3-1 bound to SARS-CoV-2 spike

EMDB-41370: 
Structure of a class A GPCR/Fab complex

EMDB-41827: 
Structure of a class A GPCR/agonist complex (focused map2)

EMDB-41828: 
Structure of a class A GPCR/agonist complex (focused map1)

EMDB-41829: 
Structure of a class A GPCR/agonist complex

EMDB-41850: 
Structure of a class A GPCR/agonist complex (Consensus map)

EMDB-41144: 
Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16

EMDB-41145: 
Cryo-EM Structure of GPR61-

EMDB-34710: 
Cryo-EM structure of WeiTsing

EMDB-40094: 
17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer

EMDB-40095: 
17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer

EMDB-13895: 
CryoEM structure of the Smc5/6-holocomplex (composite structure)

EMDB-13893: 
Cryo-EM structure of the Smc5/6 holo-complex; map for head-end of complex.

EMDB-13894: 
CryoEM structure of the Smc5/6 holocomplex; map for hinge and arm region.

EMDB-26522: 
SARS-CoV-2 6P Mut7 in complex with K398.25 Fab

EMDB-26523: 
SARS-CoV-1 in complex with K398.25 Fab

EMDB-26524: 
SARS-CoV-2 6P Mut7 in complex with K398.16 Fab
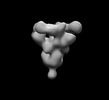
EMDB-26525: 
SARS-CoV-2 6P Mut7 in complex with K398.16 Fab (3 bound)

EMDB-26526: 
SARS-CoV-1 in complex with K398.16 Fab

EMDB-26527: 
SARS-CoV-2 6P Mut7 in complex with K288.2 Fab

EMDB-26528: 
SARS-CoV-2 6P Mut7 in complex with K398.8 Fab

EMDB-26529: 
SARS-CoV-2 6P Mut7 in complex with K398.8 Fab (2 bound)

EMDB-26530: 
SARS-CoV-2 6P Mut7 in complex with K398.18 Fab

EMDB-26531: 
SARS-CoV-2 6P Mut7 in complex with K398.18 Fabs (2 bound)

EMDB-26532: 
SARS-CoV-2 6P Mut7 in complex with K398.22 Fab
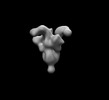
EMDB-26533: 
SARS-CoV-2 6P Mut7 in complex with K398.22 Fab (2 bound)

EMDB-26534: 
SARS-CoV-1 in complex with K398.8 Fab

EMDB-26535: 
SARS-CoV-1 in complex with K398.8 Fab (2 bound)

EMDB-26536: 
SARS-CoV-1 in complex with K398.18 Fab (2 bound)

EMDB-26537: 
SARS-CoV-1 in complex with K398.18 Fab (3 bound)

EMDB-26538: 
SARS-CoV-1 in complex with K288.2 Fab

EMDB-26539: 
SARS-CoV-1 in complex with K398.22 Fab

EMDB-12957: 
Cryo-EM structure of the human R2TP-TTT complex

EMDB-12979: 
Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex

EMDB-22943: 
Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5

EMDB-22949: 
Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5

EMDB-22950: 
Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5

EMDB-22922: 
ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4

EMDB-22927: 
Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4

EMDB-22932: 
Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4

EMDB-22941: 
Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4

EMDB-22515: 
Structure of SARS-CoV-2 spike at pH 4.5
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
