[English] 日本語
 Yorodumi
Yorodumi- EMDB-22922: ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22922 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | |||||||||
 Map data Map data | Primary sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | COVID / COVID19 / SARS-CoV2 / ACE2 / prefusion / Hydrolase-Viral Protein complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / viral translation / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction ...positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / viral translation / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / peptidyl-dipeptidase activity / regulation of vasoconstriction / transporter activator activity / Metabolism of Angiotensinogen to Angiotensins / carboxypeptidase activity / angiotensin maturation / Attachment and Entry / viral life cycle / receptor-mediated endocytosis of virus by host cell / metallocarboxypeptidase activity / positive regulation of cardiac muscle contraction / regulation of cytokine production / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / positive regulation of reactive oxygen species metabolic process / metallopeptidase activity / endocytic vesicle membrane / regulation of cell population proliferation / virus receptor activity / regulation of inflammatory response / endopeptidase activity / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / Potential therapeutics for SARS / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / apical plasma membrane / host cell surface receptor binding / cilium / symbiont-mediated suppression of host innate immune response / membrane raft / endocytosis involved in viral entry into host cell / endoplasmic reticulum lumen / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / cell surface / negative regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.39 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2020 Journal: Cell Host Microbe / Year: 2020Title: Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Authors: Tongqing Zhou / Yaroslav Tsybovsky / Jason Gorman / Micah Rapp / Gabriele Cerutti / Gwo-Yu Chuang / Phinikoula S Katsamba / Jared M Sampson / Arne Schön / Jude Bimela / Jeffrey C Boyington ...Authors: Tongqing Zhou / Yaroslav Tsybovsky / Jason Gorman / Micah Rapp / Gabriele Cerutti / Gwo-Yu Chuang / Phinikoula S Katsamba / Jared M Sampson / Arne Schön / Jude Bimela / Jeffrey C Boyington / Alexandra Nazzari / Adam S Olia / Wei Shi / Mallika Sastry / Tyler Stephens / Jonathan Stuckey / I-Ting Teng / Pengfei Wang / Shuishu Wang / Baoshan Zhang / Richard A Friesner / David D Ho / John R Mascola / Lawrence Shapiro / Peter D Kwong /  Abstract: The SARS-CoV-2 spike employs mobile receptor-binding domains (RBDs) to engage the human ACE2 receptor and to facilitate virus entry, which can occur through low-pH-endosomal pathways. To understand ...The SARS-CoV-2 spike employs mobile receptor-binding domains (RBDs) to engage the human ACE2 receptor and to facilitate virus entry, which can occur through low-pH-endosomal pathways. To understand how ACE2 binding and low pH affect spike conformation, we determined cryo-electron microscopy structures-at serological and endosomal pH-delineating spike recognition of up to three ACE2 molecules. RBDs freely adopted "up" conformations required for ACE2 interaction, primarily through RBD movement combined with smaller alterations in neighboring domains. In the absence of ACE2, single-RBD-up conformations dominated at pH 5.5, resolving into a solitary all-down conformation at lower pH. Notably, a pH-dependent refolding region (residues 824-858) at the spike-interdomain interface displayed dramatic structural rearrangements and mediated RBD positioning through coordinated movements of the entire trimer apex. These structures provide a foundation for understanding prefusion-spike mechanics governing endosomal entry; we suggest that the low pH all-down conformation potentially facilitates immune evasion from RBD-up binding antibody. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22922.map.gz emd_22922.map.gz | 267.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22922-v30.xml emd-22922-v30.xml emd-22922.xml emd-22922.xml | 26.8 KB 26.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22922.png emd_22922.png | 120.2 KB | ||
| Masks |  emd_22922_msk_1.map emd_22922_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22922.cif.gz emd-22922.cif.gz | 7.7 KB | ||
| Others |  emd_22922_additional_1.map.gz emd_22922_additional_1.map.gz emd_22922_additional_2.map.gz emd_22922_additional_2.map.gz emd_22922_additional_3.map.gz emd_22922_additional_3.map.gz emd_22922_half_map_1.map.gz emd_22922_half_map_1.map.gz emd_22922_half_map_2.map.gz emd_22922_half_map_2.map.gz | 69.9 MB 141.4 MB 264.9 MB 262.5 MB 262.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22922 http://ftp.pdbj.org/pub/emdb/structures/EMD-22922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22922 | HTTPS FTP |
-Related structure data
| Related structure data |  7kmbMC  6xluC  6xm0C  6xm3C  6xm4C  6xm5C  7jwyC  7kmsC  7kmzC  7knbC  7kneC  7knhC  7kniC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22922.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22922.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22922_msk_1.map emd_22922_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
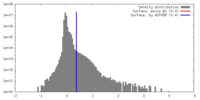
| Density Histograms |
-Additional map: C3 unmasked map
| File | emd_22922_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C3 unmasked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_22922_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened C3 unmasked map
| File | emd_22922_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened C3 unmasked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_22922_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_22922_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ACE2-RBD pH 7.4
| Entire | Name: ACE2-RBD pH 7.4 |
|---|---|
| Components |
|
-Supramolecule #1: ACE2-RBD pH 7.4
| Supramolecule | Name: ACE2-RBD pH 7.4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Angiotensin-converting enzyme 2
| Macromolecule | Name: Angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: angiotensin-converting enzyme 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.153664 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STIEEQAKTF LDKFNHEAED LFYQSSLASW NYNTNITEEN VQNMNNAGDK WSAFLKEQST LAQMYPLQEI QNLTVKLQLQ ALQQNGSSV LSEDKSKRLN TILNTMSTIY STGKVCNPDN PQECLLLEPG LNEIMANSLD YNERLWAWES WRSEVGKQLR P LYEEYVVL ...String: STIEEQAKTF LDKFNHEAED LFYQSSLASW NYNTNITEEN VQNMNNAGDK WSAFLKEQST LAQMYPLQEI QNLTVKLQLQ ALQQNGSSV LSEDKSKRLN TILNTMSTIY STGKVCNPDN PQECLLLEPG LNEIMANSLD YNERLWAWES WRSEVGKQLR P LYEEYVVL KNEMARANHY EDYGDYWRGD YEVNGVDGYD YSRGQLIEDV EHTFEEIKPL YEHLHAYVRA KLMNAYPSYI SP IGCLPAH LLGDMWGRFW TNLYSLTVPF GQKPNIDVTD AMVDQAWDAQ RIFKEAEKFF VSVGLPNMTQ GFWENSMLTD PGN VQKAVC HPTAWDLGKG DFRILMCTKV TMDDFLTAHH EMGHIQYDMA YAAQPFLLRN GANEGFHEAV GEIMSLSAAT PKHL KSIGL LSPDFQEDNE TEINFLLKQA LTIVGTLPFT YMLEKWRWMV FKGEIPKDQW MKKWWEMKRE IVGVVEPVPH DETYC DPAS LFHVSNDYSF IRYYTRTLYQ FQFQEALCQA AKHEGPLHKC DISNSTEAGQ KLFNMLRLGK SEPWTLALEN VVGAKN MNV RPLLNYFEPL FTWLKDQNKN SFVGWSTDWS PYAD UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #2: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.721266 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HAIHVSGTNG TKRFDNPVLP FNDGVYFAST EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLGVY YHKNNKSWME SEFRVYSSAN NCTFEYVSQP F LMDLEGKQ ...String: VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HAIHVSGTNG TKRFDNPVLP FNDGVYFAST EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLGVY YHKNNKSWME SEFRVYSSAN NCTFEYVSQP F LMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPINLV RDLPQGFSAL EPLVDLPIGI NITRFQTLLA LHRSYLTPGD SS SGWTAGA AAYYVGYLQP RTFLLKYNEN GTITDAVDCA LDPLSETKCT LKSFTVEKGI YQTSNFRVQP TESIVRFPNI TNL CPFGEV FNATRFASVY AWNRKRISNC VADYSVLYNS ASFSTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQT GKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSY GFQP TNGVGYQPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNGLTGT GVLTESNKKF LPFQQFGRDI ADTTDA VRD PQTLEILDIT PCSFGGVSVI TPGTNTSNQV AVLYQDVNCT EVPVAIHADQ LTPTWRVYST GSNVFQTRAG CLIGAEH VN NSYECDIPIG AGICASYQTQ TNSPGSASSV ASQSIIAYTM SLGAENSVAY SNNSIAIPTN FTISVTTEIL PVSMTKTS V DCTMYICGDS TECSNLLLQY GSFCTQLNRA LTGIAVEQDK NTQEVFAQVK QIYKTPPIKD FGGFNFSQIL PDPSKPSKR SFIEDLLFNK VTLADAGFIK QYGDCLGDIA ARDLICAQKF NGLTVLPPLL TDEMIAQYTS ALLAGTITSG WTFGAGAALQ IPFAMQMAY RFNGIGVTQN VLYENQKLIA NQFNSAIGKI QDSLSSTASA LGKLQDVVNQ NAQALNTLVK QLSSNFGAIS S VLNDILSR LDPPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QS APHGVVF LHVTYVPAQE KNFTTAPAIC HDGKAHFPRE GVFVSNGTHW FVTQRNFYEP QIITTDNTFV SGNCDVVIGI VNN TVYDPL QPELDSFKEE LDKYFKNHTS PDVDLGDISG INASVVNIQK EIDRLNEVAK NLNESLIDLQ ELGKYEQGSG YIPE APRDG QAYVRKDGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHSAW SHPQFEKGGG SGGGGSGGSA WSHPQFEK UniProtKB: Spike glycoprotein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Formula: PBS |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3104 / Average exposure time: 2.0 sec. / Average electron dose: 53.49 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-7kmb: |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)