[English] 日本語
 Yorodumi
Yorodumi- EMDB-13894: CryoEM structure of the Smc5/6 holocomplex; map for hinge and arm... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the Smc5/6 holocomplex; map for hinge and arm region. | |||||||||
 Map data Map data | Hinge and arm region of Smc5/6 holo-complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Structural Maintenance of Chromosomes / SMC / holo-complex / recombination / stalled replication fork | |||||||||
| Biological species |  | |||||||||
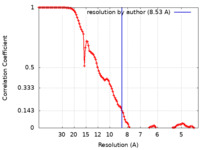
| Method | single particle reconstruction / cryo EM / Resolution: 8.53 Å | |||||||||
 Authors Authors | OLIVER AW / Hallett ST | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Cryo-EM structure of the Smc5/6 holo-complex. Authors: Stephen T Hallett / Isabella Campbell Harry / Pascale Schellenberger / Lihong Zhou / Nora B Cronin / Jonathan Baxter / Thomas J Etheridge / Johanne M Murray / Antony W Oliver /  Abstract: The Smc5/6 complex plays an essential role in the resolution of recombination intermediates formed during mitosis or meiosis, or as a result of the cellular response to replication stress. It also ...The Smc5/6 complex plays an essential role in the resolution of recombination intermediates formed during mitosis or meiosis, or as a result of the cellular response to replication stress. It also functions as a restriction factor preventing viral replication. Here, we report the cryogenic EM (cryo-EM) structure of the six-subunit budding yeast Smc5/6 holo-complex, reconstituted from recombinant proteins expressed in insect cells - providing both an architectural overview of the entire complex and an understanding of how the Nse1/3/4 subcomplex binds to the hetero-dimeric SMC protein core. In addition, we demonstrate that a region within the head domain of Smc5, equivalent to the 'W-loop' of Smc4 or 'F-loop' of Smc1, mediates an important interaction with Nse1. Notably, mutations that alter the surface-charge profile of the region of Nse1 which accepts the Smc5-loop, lead to a slow-growth phenotype and a global reduction in the chromatin-associated fraction of the Smc5/6 complex, as judged by single molecule localisation microscopy experiments in live yeast. Moreover, when taken together, our data indicates functional equivalence between the structurally unrelated KITE and HAWK accessory subunits associated with SMC complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13894.map.gz emd_13894.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13894-v30.xml emd-13894-v30.xml emd-13894.xml emd-13894.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13894_fsc.xml emd_13894_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13894.png emd_13894.png | 29.4 KB | ||
| Masks |  emd_13894_msk_1.map emd_13894_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13894.cif.gz emd-13894.cif.gz | 4.7 KB | ||
| Others |  emd_13894_half_map_1.map.gz emd_13894_half_map_1.map.gz emd_13894_half_map_2.map.gz emd_13894_half_map_2.map.gz | 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13894 http://ftp.pdbj.org/pub/emdb/structures/EMD-13894 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13894 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13894 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13894.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13894.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hinge and arm region of Smc5/6 holo-complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||
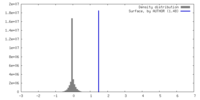
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13894_msk_1.map emd_13894_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
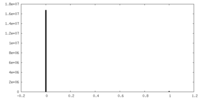
| Density Histograms |
-Half map: Half-map A for hinge and arm region of Smc5/6 holo-complex
| File | emd_13894_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A for hinge and arm region of Smc5/6 holo-complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
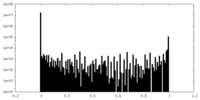
| Density Histograms |
-Half map: Half-map B for hinge and arm region of Smc5/6 holo-complex
| File | emd_13894_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B for hinge and arm region of Smc5/6 holo-complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
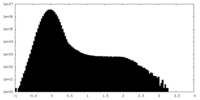
| Density Histograms |
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae Smc5/6 holo-complex
| Entire | Name: Saccharomyces cerevisiae Smc5/6 holo-complex |
|---|---|
| Components |
|
-Supramolecule #1: Saccharomyces cerevisiae Smc5/6 holo-complex
| Supramolecule | Name: Saccharomyces cerevisiae Smc5/6 holo-complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Map focussed at the hinge/arm region of the holocomplex |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR Details: Grids were glow-discharged for 60 seconds at 15 mA using a PELCO easiGlow (Agar Scientific) | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283.15 K / Instrument: LEICA EM GP Details: The grid was held in the chamber for a period of 10 seconds, before blotting for 2.5 to 4.5 seconds using the auto-sensor. Grids were then plunged into liquid ethane before being stored ...Details: The grid was held in the chamber for a period of 10 seconds, before blotting for 2.5 to 4.5 seconds using the auto-sensor. Grids were then plunged into liquid ethane before being stored under liquid nitrogen until data collection.. | ||||||||||||
| Details | Fraction(s) eluting from a Superose 6 size exclusion chromatography column (Cytiva Life Sciences). Cross-linked with BS3 before grid preparation. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 4 / Average exposure time: 4.0 sec. / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)