-Search query
-Search result
Showing all 43 items for (author: trono & d)

EMDB-17819: 
XBB 1.0 RBD bound to P4J15 (Local)
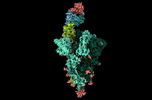
EMDB-17849: 
XBB 1.0 RBD bound to P4J15 (Global)

EMDB-17850: 
SARS-CoV-2 XBB 1.0 closed conformation.

PDB-8pq2: 
XBB 1.0 RBD bound to P4J15 (Local)

PDB-8psd: 
SARS-CoV-2 XBB 1.0 closed conformation.
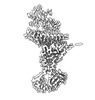
EMDB-15592: 
BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqw: 
BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local)

EMDB-14922: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map

PDB-7zrv: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map

EMDB-14930: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local
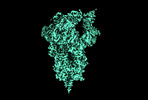
EMDB-14947: 
cryo-EM structure of D614 spike in complex with de novo designed binder, full and local maps(addition)

PDB-7zsd: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local

PDB-7zss: 
cryo-EM structure of D614 spike in complex with de novo designed binder
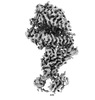
EMDB-15588: 
BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local)

EMDB-15589: 
Beta SARS-CoV-2 Spike bound to mouse ACE2 (local)

EMDB-15590: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)
EMDB-15591: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqs: 
BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local)

PDB-8aqt: 
Beta SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqu: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqv: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)

EMDB-15541: 
Beta SARS-CoV-2 Spike bound to mouse ACE2 (two up, full)
EMDB-15580: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (two up, full)
EMDB-15581: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (three up, full)
EMDB-15584: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (two up, full)
EMDB-15585: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (three up, full)

EMDB-15586: 
BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (two up, full)

EMDB-15587: 
BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (three up, full)

EMDB-14141: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)

EMDB-14142: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)
EMDB-14143: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local)

PDB-7qti: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)

PDB-7qtj: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)

PDB-7qtk: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local)

EMDB-14087: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD and NTD (Local)

PDB-7qo9: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD and NTD (Local)

EMDB-14086: 
SARS-CoV-2 S Omicron Spike B.1.1.529

PDB-7qo7: 
SARS-CoV-2 S Omicron Spike B.1.1.529

EMDB-13190: 
P5C3 is a potent fab neutralizer

EMDB-13265: 
the local resolution of Fab p5c3.

EMDB-13415: 
MaP OF P5C3RBD Interface

PDB-7p40: 
P5C3 is a potent fab neutralizer

PDB-7phg: 
MaP OF P5C3RBD Interface
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
