[English] 日本語
 Yorodumi
Yorodumi- EMDB-14141: SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 S Omicron Spike B.1.1.529 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Ni D / Lau K / Turelli P / Fenwick C / Perez L / Pojer F / Stahlberg H / Pantaleo G / Trono D | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2022 Journal: Nat Microbiol / Year: 2022Title: Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys. Authors: Craig Fenwick / Priscilla Turelli / Dongchun Ni / Laurent Perez / Kelvin Lau / Cécile Herate / Romain Marlin / Erica Lana / Céline Pellaton / Charlène Raclot / Line Esteves-Leuenberger / ...Authors: Craig Fenwick / Priscilla Turelli / Dongchun Ni / Laurent Perez / Kelvin Lau / Cécile Herate / Romain Marlin / Erica Lana / Céline Pellaton / Charlène Raclot / Line Esteves-Leuenberger / Jérémy Campos / Alex Farina / Flurin Fiscalini / Nathalie Dereuddre-Bosquet / Francis Relouzat / Rana Abdelnabi / Caroline S Foo / Johan Neyts / Pieter Leyssen / Yves Lévy / Florence Pojer / Henning Stahlberg / Roger LeGrand / Didier Trono / Giuseppe Pantaleo /    Abstract: The SARS-CoV-2 Omicron variant has very high levels of transmission, is resistant to neutralization by authorized therapeutic human monoclonal antibodies (mAb) and is less sensitive to vaccine- ...The SARS-CoV-2 Omicron variant has very high levels of transmission, is resistant to neutralization by authorized therapeutic human monoclonal antibodies (mAb) and is less sensitive to vaccine-mediated immunity. To provide additional therapies against Omicron, we isolated a mAb named P2G3 from a previously infected vaccinated donor and showed that it has picomolar-range neutralizing activity against Omicron BA.1, BA.1.1, BA.2 and all other variants tested. We solved the structure of P2G3 Fab in complex with the Omicron spike using cryo-electron microscopy at 3.04 Å resolution to identify the P2G3 epitope as a Class 3 mAb that is different from mAb-binding spike epitopes reported previously. Using a SARS-CoV-2 Omicron monkey challenge model, we show that P2G3 alone, or in combination with P5C3 (a broadly active Class 1 mAb previously identified), confers complete prophylactic or therapeutic protection. Although we could select for SARS-CoV-2 mutants escaping neutralization by P2G3 or by P5C3 in vitro, they had low infectivity and 'escape' mutations are extremely rare in public sequence databases. We conclude that this combination of mAbs has potential as an anti-Omicron drug. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14141.map.gz emd_14141.map.gz | 97.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14141-v30.xml emd-14141-v30.xml emd-14141.xml emd-14141.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14141_fsc.xml emd_14141_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14141.png emd_14141.png | 61.8 KB | ||
| Filedesc metadata |  emd-14141.cif.gz emd-14141.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14141 http://ftp.pdbj.org/pub/emdb/structures/EMD-14141 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14141 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14141 | HTTPS FTP |
-Related structure data
| Related structure data |  7qtiMC  7qtjC  7qtkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14141.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14141.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38333 Å | ||||||||||||||||||||||||||||||||||||
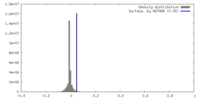
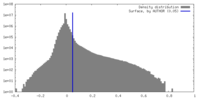
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Spike Omicron trimer with Fabs
| Entire | Name: Spike Omicron trimer with Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Spike Omicron trimer with Fabs
| Supramolecule | Name: Spike Omicron trimer with Fabs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: Omicron Spike
| Supramolecule | Name: Omicron Spike / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Fabs
| Supramolecule | Name: Fabs / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: P2G3 Heavy Chain
| Macromolecule | Name: P2G3 Heavy Chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.562475 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGRSLRL SCAASGFRFD DYALHWVRQA PEKGLEWVSG ISWNSNNIGY AESVKGRFTI SRDTAKKSLY LQMNDLRAE DTALYYCVKD RHYDSSGYFV NGFDIWGQGT LVTVSAASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS ...String: EVQLVESGGG LVQPGRSLRL SCAASGFRFD DYALHWVRQA PEKGLEWVSG ISWNSNNIGY AESVKGRFTI SRDTAKKSLY LQMNDLRAE DTALYYCVKD RHYDSSGYFV NGFDIWGQGT LVTVSAASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC |
-Macromolecule #2: P2G3 Light Chain
| Macromolecule | Name: P2G3 Light Chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.321771 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQLTQSPSF LSASVGDRVT VTCRASQGIS SYVAWYQQKA GKAPTLLIYT ASTLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQ QLHSYPVTFG QGTRLDIERT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQLTQSPSF LSASVGDRVT VTCRASQGIS SYVAWYQQKA GKAPTLLIYT ASTLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQ QLHSYPVTFG QGTRLDIERT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: P5C3 Light Chain
| Macromolecule | Name: P5C3 Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.388875 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRGSQSVR SSYLGWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGSSPWTF GQGTKVEIKG TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGERAT LSCRGSQSVR SSYLGWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGSSPWTF GQGTKVEIKG TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: P5C3 Heavy Chain
| Macromolecule | Name: P5C3 Heavy Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.362133 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QMQLVQSGPE VKKPGTSVKV SCKASGFTFT SSAVQWVRQA RGQRLEWIGW IVVGSGNTDY AQQFQERVTI TRDMSTSTAY MELSSLGSE DTAVYYCAAP NCSGGSCYDG FDLWGQGTMV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QMQLVQSGPE VKKPGTSVKV SCKASGFTFT SSAVQWVRQA RGQRLEWIGW IVVGSGNTDY AQQFQERVTI TRDMSTSTAY MELSSLGSE DTAVYYCAAP NCSGGSCYDG FDLWGQGTMV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKV |
-Macromolecule #5: Spike glycoprotein,Envelope glycoprotein
| Macromolecule | Name: Spike glycoprotein,Envelope glycoprotein / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 142.558094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ PFLMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPIIVR EPEDLPQGFS ALEPLVDLPI GINITRFQTL LA LHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFD EVFNATRFAS VYAWNRKRIS NCVADYSVLY NLAPFFTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKVSGN YNYLYRLFRK SNLKPFERDI STEIYQAGNK PCNGV AGFN CYFPLRSYSF RPTYGVGHQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLK GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEY VNNSYECDIP IGAGICASYQ TQTKSHGSAS SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLK RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KYFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFKGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTA SALGKLQDVV NHNAQALNTL V KQLSSKFG AISSVLNDIF SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG RSLEVLFQGP GHHHHHHHHS AWSHPQFEKG GGSGGGGSGG SAWSH PQFE K UniProtKB: Spike glycoprotein, Envelope glycoprotein |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 26 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: PBS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)