-Search query
-Search result
Showing all 34 items for (author: sinha & d)
EMDB-29837: 
Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of nucleosome)
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M
EMDB-29841: 
Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of Oct4 bound region)
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M
EMDB-29843: 
Human Oct4 bound to nucleosome with human nMatn1 sequence
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M
EMDB-29845: 
Nucleosome with human nMatn1 sequence in complex with Human Oct4
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M
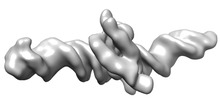
EMDB-29846: 
Human Oct4 bound to nucleosome with human LIN28B sequence
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M
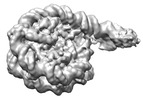
EMDB-29850: 
Interaction of H3 tail in LIN28B nucleosome with Oct4
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

EMDB-29852: 
Oct4 interaction with H2A C-terminal tail in LIN28B nucleosome
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

EMDB-29853: 
H4 tail conformation A in Oct4 bound LIN28 nucleosome
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M
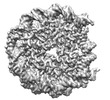
EMDB-29854: 
H4 tail conformation B in Oct4 bound LIN28B nucleosome
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M
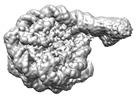
EMDB-29855: 
All Oct4 bound LIN28 nucleosomes
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

PDB-8g86: 
Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of nucleosome)
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

PDB-8g87: 
Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of Oct4 bound region)
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

PDB-8g88: 
Human Oct4 bound to nucleosome with human nMatn1 sequence
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

PDB-8g8b: 
Nucleosome with human nMatn1 sequence in complex with Human Oct4
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

PDB-8g8e: 
Human Oct4 bound to nucleosome with human LIN28B sequence
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

PDB-8g8g: 
Interaction of H3 tail in LIN28B nucleosome with Oct4
Method: single particle / : Sinha KK, Bilokapic S, Du Y, Malik D, Halic M

EMDB-25498: 
CV3-25 IgG in complex with SARS-CoV-2 6P-D614G S protein
Method: single particle / : Jackson AM, Ozorowski G, Ward AB

EMDB-12882: 
A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ)
Method: single particle / : Dendooven T, Sinha D

EMDB-12883: 
A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. apo-PNPase
Method: single particle / : Dendooven T, Sinha D

EMDB-12884: 
A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ)-Hfq
Method: single particle / : Dendooven T, Sinha D

PDB-7ogk: 
A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ)
Method: single particle / : Dendooven T, Sinha D, Roesoleva A, Cameron TA, De Lay N, Luisi BF, Bandyra K

PDB-7ogl: 
A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. apo-PNPase
Method: single particle / : Dendooven T, Sinha D, Roesoleva A, Cameron TA, De Lay N, Luisi BF, Bandyra K

PDB-7ogm: 
A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ)-Hfq
Method: single particle / : Dendooven T, Sinha D, Roesoleva A, Cameron TA, De Lay N, Luisi BF, Bandyra K

EMDB-11457: 
Mbf1-ribosome complex
Method: single particle / : Best KM, Denk T

PDB-6zvi: 
Mbf1-ribosome complex
Method: single particle / : Best KM, Denk T, Cheng J, Thoms M, Berninghausen O, Beckmann R

EMDB-11456: 
EDF1-ribosome complex
Method: single particle / : Best KM, Denk T

PDB-6zvh: 
EDF1-ribosome complex
Method: single particle / : Best KM, Denk T, Cheng J, Thoms M, Berninghausen O, Beckmann R

EMDB-0103: 
Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses
Method: single particle / : Geraets JA, Flatt JW

PDB-6gzv: 
Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses
Method: single particle / : Geraets JA, Flatt JW, Domanska A, Butcher SJ

EMDB-7292: 
Drosophila Dicer-2 (apo, intact helicase density)
Method: single particle / : Shen PS, Sinha NK, Bass BL

EMDB-7291: 
Drosophila Dicer-2 apo homology model (helicase, Platform-PAZ, RNaseIII domains)
Method: single particle / : Shen PS, Sinha NK
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
