[English] 日本語
 Yorodumi
Yorodumi- EMDB-0103: Identification of a druggable VP1-VP3 interprotomer pocket in the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0103 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses | |||||||||
 Map data Map data | Coxsackievirus B3 Nancy in complex with a small-molecule inhibitor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibitor / complex / virus | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host transcription / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane ...symbiont-mediated perturbation of host transcription / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / symbiont-mediated suppression of host NF-kappaB cascade / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Coxsackievirus B3 (strain Nancy) Coxsackievirus B3 (strain Nancy) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Geraets JA / Flatt JW | |||||||||
| Funding support |  Finland, 1 items Finland, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2019 Journal: PLoS Biol / Year: 2019Title: A novel druggable interprotomer pocket in the capsid of rhino- and enteroviruses. Authors: Rana Abdelnabi / James A Geraets / Yipeng Ma / Carmen Mirabelli / Justin W Flatt / Aušra Domanska / Leen Delang / Dirk Jochmans / Timiri Ajay Kumar / Venkatesan Jayaprakash / Barij Nayan ...Authors: Rana Abdelnabi / James A Geraets / Yipeng Ma / Carmen Mirabelli / Justin W Flatt / Aušra Domanska / Leen Delang / Dirk Jochmans / Timiri Ajay Kumar / Venkatesan Jayaprakash / Barij Nayan Sinha / Pieter Leyssen / Sarah J Butcher / Johan Neyts /    Abstract: Rhino- and enteroviruses are important human pathogens, against which no antivirals are available. The best-studied inhibitors are "capsid binders" that fit in a hydrophobic pocket of the viral ...Rhino- and enteroviruses are important human pathogens, against which no antivirals are available. The best-studied inhibitors are "capsid binders" that fit in a hydrophobic pocket of the viral capsid. Employing a new class of entero-/rhinovirus inhibitors and by means of cryo-electron microscopy (EM), followed by resistance selection and reverse genetics, we discovered a hitherto unknown druggable pocket that is formed by viral proteins VP1 and VP3 and that is conserved across entero-/rhinovirus species. We propose that these inhibitors stabilize a key region of the virion, thereby preventing the conformational expansion needed for viral RNA release. A medicinal chemistry effort resulted in the identification of analogues targeting this pocket with broad-spectrum activity against Coxsackieviruses B (CVBs) and compounds with activity against enteroviruses (EV) of groups C and D, and even rhinoviruses (RV). Our findings provide novel insights in the biology of the entry of entero-/rhinoviruses and open new avenues for the design of broad-spectrum antivirals against these pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0103.map.gz emd_0103.map.gz | 55.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0103-v30.xml emd-0103-v30.xml emd-0103.xml emd-0103.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0103_fsc.xml emd_0103_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_0103.png emd_0103.png | 291.5 KB | ||
| Filedesc metadata |  emd-0103.cif.gz emd-0103.cif.gz | 6.6 KB | ||
| Others |  emd_0103_half_map_1.map.gz emd_0103_half_map_1.map.gz emd_0103_half_map_2.map.gz emd_0103_half_map_2.map.gz | 128.2 MB 128.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0103 http://ftp.pdbj.org/pub/emdb/structures/EMD-0103 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0103 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0103 | HTTPS FTP |
-Related structure data
| Related structure data |  6gzvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10199 (Title: Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses EMPIAR-10199 (Title: Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enterovirusesData size: 2.2 TB Data #1: Unaligned multi-frame micrographs of CVB3 in complex with a small molecular inhibitor [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0103.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0103.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Coxsackievirus B3 Nancy in complex with a small-molecule inhibitor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_0103_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_0103_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Coxsackievirus B3 (strain Nancy)
| Entire | Name:  Coxsackievirus B3 (strain Nancy) Coxsackievirus B3 (strain Nancy) |
|---|---|
| Components |
|
-Supramolecule #1: Coxsackievirus B3 (strain Nancy)
| Supramolecule | Name: Coxsackievirus B3 (strain Nancy) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Coxsackievirus B3 (strain Nancy) NCBI Taxonomy ID: 103903 Full, non-enveloped virion from natural source Diameter: ~314A Triangulation: 1 (pT3) Benzene sulfonamide derivative Molecular ...Details: Coxsackievirus B3 (strain Nancy) NCBI Taxonomy ID: 103903 Full, non-enveloped virion from natural source Diameter: ~314A Triangulation: 1 (pT3) Benzene sulfonamide derivative Molecular weight: 422Da Binding sites on capsid: 60 NCBI-ID: 103903 / Sci species name: Coxsackievirus B3 (strain Nancy) / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Diameter: 314.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B3 (strain Nancy) / Strain: Nancy Coxsackievirus B3 (strain Nancy) / Strain: Nancy |
| Molecular weight | Theoretical: 31.639438 KDa |
| Sequence | String: GPVEDAITAA IGRVADTVGT GPTNSEAIPA LTAAETGHTS QVVPGDTMQT RHVKNYHSRS ESTIENFLCR SACVYFTEYK NSGAKRYAE WVLTPRQAAQ LRRKLEFFTY VRFDLELTFV ITSTQQPSTT QNQDAQILTH QIMYVPPGGP VPDKVDSYVW Q TSTNPSVF ...String: GPVEDAITAA IGRVADTVGT GPTNSEAIPA LTAAETGHTS QVVPGDTMQT RHVKNYHSRS ESTIENFLCR SACVYFTEYK NSGAKRYAE WVLTPRQAAQ LRRKLEFFTY VRFDLELTFV ITSTQQPSTT QNQDAQILTH QIMYVPPGGP VPDKVDSYVW Q TSTNPSVF WTEGNAPPRM SIPFLSIGNA YSNFYDGWSE FSRNGVYGIN TLNNMGTLYA RHVNAGSTGP IKSTIRIYFK PK HVKAWIP RPPRLCQYEK AKNVNFQPSG VTTTRQSITT MTNTGAF UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B3 (strain Nancy) / Strain: Nancy Coxsackievirus B3 (strain Nancy) / Strain: Nancy |
| Molecular weight | Theoretical: 28.836502 KDa |
| Sequence | String: SPTVEECGYS DRARSITLGN STITTQECAN VVVGYGVWPD YLKDSEATAE DQPTQPDVAT CRFYTLDSVQ WQKTSPGWWW KLPDALSNL GLFGQNMQYH YLGRTGYTVH VQCNASKFHQ GCLLVVCVPE AEMGCATLDN TPSSAELLGG DTAKEFADKP V ASGSNKLV ...String: SPTVEECGYS DRARSITLGN STITTQECAN VVVGYGVWPD YLKDSEATAE DQPTQPDVAT CRFYTLDSVQ WQKTSPGWWW KLPDALSNL GLFGQNMQYH YLGRTGYTVH VQCNASKFHQ GCLLVVCVPE AEMGCATLDN TPSSAELLGG DTAKEFADKP V ASGSNKLV QRVVYNAGMG VGVGNLTIFP HQWINLRTNN SATIVMPYTN SVPMDNMFRH NNVTLMVIPF VPLDYCPGST TY VPITVTI APMCAEYNGL RLAGHQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B3 (strain Nancy) / Strain: Nancy Coxsackievirus B3 (strain Nancy) / Strain: Nancy |
| Molecular weight | Theoretical: 26.195727 KDa |
| Sequence | String: GLPTMNTPGS CQFLTSDDFQ SPSAMPQYDV TPEMRIPGEV KNLMEIAEVD SVVPVQNVGE KVNSMEAYQI PVRSNEGSGT QVFGFPLQP GYSSVFSRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GAPTKRVDAM LGTHVIWDVG L QSSCVLCI ...String: GLPTMNTPGS CQFLTSDDFQ SPSAMPQYDV TPEMRIPGEV KNLMEIAEVD SVVPVQNVGE KVNSMEAYQI PVRSNEGSGT QVFGFPLQP GYSSVFSRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GAPTKRVDAM LGTHVIWDVG L QSSCVLCI PWISQTHYRF VASDEYTAGG FITCWYQTNI VVPADAQSSC YIMCFVSACN DFSVRLLKDT PFISQQNFFQ UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B3 (strain Nancy) / Strain: Nancy Coxsackievirus B3 (strain Nancy) / Strain: Nancy |
| Molecular weight | Theoretical: 7.580377 KDa |
| Sequence | String: MGAQVSTQKT GAHETRLNAS GNSIIHYTNI NYYKDAASNS ANRQDFTQDP GKFTEPVKDI MIKSLPALN UniProtKB: Genome polyprotein |
-Macromolecule #5: 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]b...
| Macromolecule | Name: 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid type: ligand / ID: 5 / Number of copies: 1 / Formula: FHK |
|---|---|
| Molecular weight | Theoretical: 422.411 Da |
| Chemical component information | 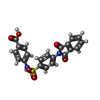 ChemComp-FHK: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER | |||||||||
| Details | Inhibitor was added at 2500:1 molar ratio to purified CVB3 and incubated for 1h at 37C |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6gzv: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































