-Search query
-Search result
Showing 1 - 50 of 165 items for (author: sauer & m)

EMDB-17377: 
Structure of human SIT1 (focussed map / refinement)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

EMDB-17378: 
Structure of human SIT1:ACE2 complex (open PD conformation)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

EMDB-17379: 
Structure of human SIT1:ACE2 complex (closed PD conformation)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

EMDB-17380: 
Structure of human SIT1 bound to L-pipecolate (focussed map / refinement)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

EMDB-17381: 
Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

EMDB-17382: 
Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

PDB-8p2w: 
Structure of human SIT1 (focussed map / refinement)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

PDB-8p2x: 
Structure of human SIT1:ACE2 complex (open PD conformation)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

PDB-8p2y: 
Structure of human SIT1:ACE2 complex (closed PD conformation)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

PDB-8p2z: 
Structure of human SIT1 bound to L-pipecolate (focussed map / refinement)
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

PDB-8p30: 
Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

PDB-8p31: 
Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate
Method: single particle / : Li HZ, Pike ACW, Chi G, Hansen JS, Lee SG, Rodstrom KEJ, Bushell SR, Speedman D, Evans A, Wang D, He D, Shrestha L, Nasrallah C, Chalk R, Moreira T, MacLean EM, Marsden B, Bountra C, Burgess-Brown NA, Dafforn TR, Carpenter EP, Sauer DB

EMDB-19109: 
Structure of XPD stalled at a Y-fork DNA containing a interstrand crosslink
Method: single particle / : Kuper J, Hove T, Kisker C

PDB-8rev: 
Structure of XPD stalled at a Y-fork DNA containing a interstrand crosslink
Method: single particle / : Kuper J, Hove T, Kisker C
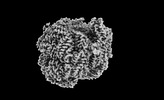
EMDB-41434: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA
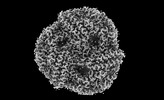
EMDB-41435: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA
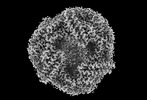
EMDB-41436: 
Central rod disk in D3 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA
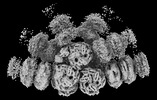
EMDB-41463: 
Synechocystis PCC 6803 Phycobilisome quenched by OCP, high resolution
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB-41475: 
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L
EMDB-41585: 
Rod from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to2: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to5: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8tpj: 
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8tro: 
Rod from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L
EMDB-40572: 
Human adenylyl Cyclase 5 in complex with Gbg
Method: single particle / : Yen YC, Tesmer JJG

EMDB-40573: 
Dimeric form of human adenylyl cyclase 5
Method: single particle / : Yen YC, Tesmer JJG

PDB-8sl3: 
Human adenylyl Cyclase 5 in complex with Gbg
Method: single particle / : Yen YC, Tesmer JJG

PDB-8sl4: 
Dimeric form of human adenylyl cyclase 5
Method: single particle / : Yen YC, Tesmer JJG

EMDB-18659: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Pilati N, Marasco A, Gunthorpe M, Alvaro G, Large C, Lakshminaraya B, Williams E, Sauer DB

EMDB-18660: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Lakshminarayana B, Pilati N, Marasco A, Gunthorpe M, Alvaro GS, Large CH, Williams E, Sauer DB

PDB-8quc: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Pilati N, Marasco A, Gunthorpe M, Alvaro G, Large C, Lakshminaraya B, Williams E, Sauer DB

PDB-8qud: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Lakshminarayana B, Pilati N, Marasco A, Gunthorpe M, Alvaro GS, Large CH, Williams E, Sauer DB

EMDB-18387: 
FtsH1 protease from P.aeruginosa clone C in negative stain
Method: single particle / : Mawla GD, Mansour Kamal S, Cao LY, Purhonen P, Hebert H, Sauer RT, Baker TA, Romling U

EMDB-18388: 
FtsH2 protease from P.aeruginosa clone C in negative stain
Method: single particle / : Mawla GD, Mansour Kamal S, Cao LY, Purhonen P, Hebert H, Sauer RT, Baker TA, Romling U

EMDB-18389: 
P.aeruginosa clone C construct PaFtsH2-H1-link32 in negative stain
Method: single particle / : Mawla GD, Mansour Kamal S, Cao LY, Purhonen P, Hebert H, Sauer RT, Baker TA, Romling U

EMDB-33329: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/+ Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

EMDB-33330: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/- Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

PDB-7xnx: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/+ Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

PDB-7xny: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/- Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

EMDB-26554: 
ClpAP complex bound to ClpS N-terminal extension, class IIa
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

EMDB-26555: 
ClpAP complex bound to ClpS N-terminal extension, class IIb
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

EMDB-26556: 
ClpAP complex bound to ClpS N-terminal extension, class I
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

EMDB-26558: 
ClpAP complex bound to ClpS N-terminal extension, class IIc
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

EMDB-26559: 
ClpAP complex bound to ClpS N-terminal extension, class IIIb
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

PDB-7uiv: 
ClpAP complex bound to ClpS N-terminal extension, class IIa
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

PDB-7uiw: 
ClpAP complex bound to ClpS N-terminal extension, class IIb
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

PDB-7uix: 
ClpAP complex bound to ClpS N-terminal extension, class I
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

PDB-7uiz: 
ClpAP complex bound to ClpS N-terminal extension, class IIc
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

PDB-7uj0: 
ClpAP complex bound to ClpS N-terminal extension, class IIIb
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA

EMDB-26557: 
ClpAP complex bound to ClpS N-terminal extension, class IIIa
Method: single particle / : Kim S, Fei X, Sauer RT, Baker TA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
