-Search query
-Search result
Showing 1 - 50 of 99 items for (author: ooi & sa)

EMDB-18887: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP)

PDB-8r4i: 
Cryo-EM structure of human islet amyloid polypeptide (hIAPP)

EMDB-36480: 
CryoEM structure of isNS1 in complex with Fab56.2 and HDL

EMDB-36483: 
CryoEM structure of isNS1 in complex with Fab56.2

EMDB-32839: 
CryoEM structure of sNS1 complexed with Fab5E3

EMDB-32840: 
CryoEM structure of a dimer of loose sNS1 tetramer

EMDB-32841: 
CryoEM structure of stable sNS1 tetramer

EMDB-32842: 
CryoEM structure of loose sNS1 tetramer

EMDB-32843: 
CryoEM structure of sNS1 hexamer

PDB-7wur: 
CryoEM structure of sNS1 complexed with Fab5E3

PDB-7wus: 
CryoEM structure of a dimer of loose sNS1 tetramer

PDB-7wut: 
CryoEM structure of stable sNS1 tetramer

PDB-7wuu: 
CryoEM structure of loose sNS1 tetramer

PDB-7wuv: 
CryoEM structure of sNS1 hexamer
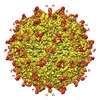
EMDB-25606: 
Zika Virus particle bound with IgM antibody DH1017 Fab fragment

PDB-7t17: 
Zika Virus asymmetric unit bound with IgM antibody DH1017 Fab fragment
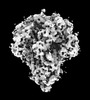
EMDB-32329: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.

EMDB-32332: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.

EMDB-32333: 
Subtomogram averaging of PEDV (Pintung 52) S protein with one protomer in the D0-up conformation and two protomers in the D0-down conformation, determined in situ on intact viral particles

EMDB-32337: 
Subtomogram averaging of PEDV (Pintung 52) S protein with two protomers in the D0-up conformation and one protomer in the D0-down conformation, determined in situ on intact viral particles.

EMDB-32338: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation
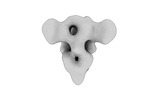
EMDB-32339: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-up conformation determined in situ on intact viral particles.

EMDB-32340: 
Subtomogram averaging of PEDV (Pintung 52) S protein in the postfusion form determined in situ on intact viral particles.

EMDB-33646: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up

EMDB-33647: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up

EMDB-33648: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation
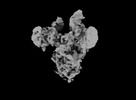
EMDB-33649: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation

EMDB-33700: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down
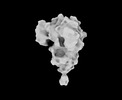
EMDB-33701: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein one D0-up and two D0-down

EMDB-33702: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-up
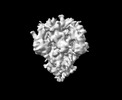
EMDB-33703: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-down

EMDB-33704: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up and two D0-down
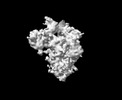
EMDB-33705: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-down and two D0-up
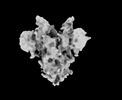
EMDB-33706: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-up

PDB-7w6m: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.

PDB-7w73: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation

PDB-7y6s: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up

PDB-7y6t: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up

PDB-7y6u: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation

PDB-7y6v: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation

EMDB-21992: 
GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor

EMDB-21993: 
Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor

EMDB-21994: 
Non peptide agonist PF-06882961, bound to Glucagon-Like peptide-1 (GLP-1) Receptor

PDB-6x18: 
GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor

PDB-6x19: 
Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor

PDB-6x1a: 
Non peptide agonist PF-06882961, bound to Glucagon-Like peptide-1 (GLP-1) Receptor

EMDB-10239: 
Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11)

EMDB-10240: 
Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,11)

EMDB-10246: 
Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,12)

PDB-6slq: 
Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
