-Search query
-Search result
Showing all 37 items for (author: noble & me)

EMDB-19408: 
Cryo-EM structure of CDK2-cyclin A in complex with CDC25A

EMDB-16325: 
Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex
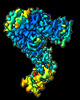
EMDB-16327: 
Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex
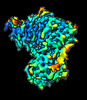
EMDB-16344: 
Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex

PDB-8bya: 
Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex

PDB-8byl: 
Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex

PDB-8bzo: 
Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex

EMDB-22078: 
Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis

PDB-6x6p: 
Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis

EMDB-0553: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2

PDB-6nz0: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2

EMDB-0621: 
Structure of the AAV2 with its Cell Receptor, AAVR
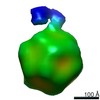
EMDB-0622: 
Structure of the AAV2 with its Cell Receptor, AAVR

EMDB-0623: 
Structure of the AAV2 with its Cell Receptor, AAVR

EMDB-0624: 
Structure of the AAV2 with its Cell Receptor, AAVR

EMDB-7076: 
Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex

PDB-6bbm: 
Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex

EMDB-7135: 
Hemagglutinin on a carbon nanowire grid plunged with Spotiton

EMDB-7138: 
Rabbit muscle aldolase on a gold nanowire grid plunged with Spotiton

EMDB-7139: 
Rabbit muscle aldolase on a carbon nanowire grid plunged with Spotiton

EMDB-7140: 
Protein in nanodisc on a gold nanowire grid plunged with Spotiton

EMDB-7141: 
Glutamate dehydrogenase on a holey carbon grid

EMDB-7142: 
Glutamate dehydrogenase on a holey carbon grid

EMDB-7143: 
GDH + 0.001% DDM on a carbon nanowire grid plunged with Spotiton

EMDB-7144: 
DNAB helicase-helicase loader on a gold Quantifoil grid

EMDB-7145: 
Apoferritin on a gold nanowire grid plunged with Spotiton

EMDB-7146: 
Apoferritin on a gold nanowire grid plunged with Spotiton

EMDB-7147: 
Apoferritin on a holey carbon nanowire grid plunged with Spotiton

EMDB-7148: 
Apoferritin on a holey carbon nanowire grid plunged with Spotiton

EMDB-7149: 
Apoferritin on a holey gold nanowire grid plunged with Spotiton

EMDB-7150: 
Apoferritin with 0.5 mM TCEP on a carbon nanowire grid plunged with Spotiton

EMDB-7151: 
T20S proteasome on a holey carbon grid

EMDB-7152: 
T20S proteasome on a holey carbon grid

EMDB-7153: 
T20S proteasome on a gold Quantifoil grid

EMDB-7154: 
Mtb 20S proteasome on a carbon nanowire grid plunged with Spotiton

EMDB-8574: 
Single particle reconstruction of chimeric adeno-associated virus-DJ with a Heparanoid Pentasaccharide

PDB-5uf6: 
The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
