[English] 日本語
 Yorodumi
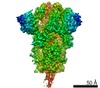
Yorodumi- PDB-6x6p: Characterization of the SARS-CoV-2 S Protein: Biophysical, Bioche... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6x6p | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | |||||||||
 Components Components | Spike glycoprotein | |||||||||
 Keywords Keywords | VIRAL PROTEIN / coronavirus / SARS-CoV-2 / SARS-CoV / spike glycoprotein / fusion protein / trimer | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.22 Å | |||||||||
 Authors Authors | Herrera, N.G. / Morano, N.C. / Celikgil, A. / Georgiev, G.I. / Malonis, R. / Lee, J.H. / Tong, K. / Vergnolle, O. / Massimi, A. / Yen, L.Y. ...Herrera, N.G. / Morano, N.C. / Celikgil, A. / Georgiev, G.I. / Malonis, R. / Lee, J.H. / Tong, K. / Vergnolle, O. / Massimi, A. / Yen, L.Y. / Noble, A.J. / Kopylov, M. / Bonanno, J.B. / Garrett-Thompson, S.C. / Hayes, D.B. / Brenowitz, M. / Garforth, S.J. / Eng, E.T. / Lai, J.R. / Almo, S.C. | |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis. Authors: Natalia G Herrera / Nicholas C Morano / Alev Celikgil / George I Georgiev / Ryan J Malonis / James H Lee / Karen Tong / Olivia Vergnolle / Aldo B Massimi / Laura Y Yen / Alex J Noble / ...Authors: Natalia G Herrera / Nicholas C Morano / Alev Celikgil / George I Georgiev / Ryan J Malonis / James H Lee / Karen Tong / Olivia Vergnolle / Aldo B Massimi / Laura Y Yen / Alex J Noble / Mykhailo Kopylov / Jeffrey B Bonanno / Sarah C Garrett-Thomson / David B Hayes / Robert H Bortz / Ariel S Wirchnianski / Catalina Florez / Ethan Laudermilch / Denise Haslwanter / J Maximilian Fels / M Eugenia Dieterle / Rohit K Jangra / Jason Barnhill / Amanda Mengotto / Duncan Kimmel / Johanna P Daily / Liise-Anne Pirofski / Kartik Chandran / Michael Brenowitz / Scott J Garforth / Edward T Eng / Jonathan R Lai / Steven C Almo /  Abstract: Coronavirus disease 2019 ( ) is a global health crisis caused by the novel severe acute respiratory syndrome coronavirus 2 ( ), and there is a critical need to produce large quantities of high- ...Coronavirus disease 2019 ( ) is a global health crisis caused by the novel severe acute respiratory syndrome coronavirus 2 ( ), and there is a critical need to produce large quantities of high-quality SARS-CoV-2 Spike ( ) protein for use in both clinical and basic science settings. To address this need, we have evaluated the expression and purification of two previously reported S protein constructs in Expi293F and ExpiCHO-S cells, two different cell lines selected for increased expression of secreted glycoproteins. We show that ExpiCHO-S cells produce enhanced yields of both SARS-CoV-2 S proteins. Biochemical, biophysical, and structural ( ) characterization of the SARS-CoV-2 S proteins produced in both cell lines demonstrate that the reported purification strategy yields high quality S protein (non-aggregated, uniform material with appropriate biochemical and biophysical properties). Importantly, we show that multiple preparations of these two recombinant S proteins from either cell line exhibit identical behavior in two different serology assays. We also evaluate the specificity of S protein-mediated host cell binding by examining interactions with proposed binding partners in the human secretome. In addition, the antigenicity of these proteins is demonstrated by standard ELISAs, and in a flexible protein microarray format. Collectively, we establish an array of metrics for ensuring the production of high-quality S protein to support clinical, biological, biochemical, structural and mechanistic studies to combat the global pandemic caused by SARS-CoV-2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6x6p.cif.gz 6x6p.cif.gz | 583.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6x6p.ent.gz pdb6x6p.ent.gz | 452.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6x6p.json.gz 6x6p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/x6/6x6p https://data.pdbj.org/pub/pdb/validation_reports/x6/6x6p ftp://data.pdbj.org/pub/pdb/validation_reports/x6/6x6p ftp://data.pdbj.org/pub/pdb/validation_reports/x6/6x6p | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  22078MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10433 (Title: Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis EMPIAR-10433 (Title: Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic AnalysisData size: 484.3 Data #1: Unaligned multi frame micrographs of SARS CoV-2 SPIKE protein produced in CHO cells [micrographs - multiframe] Data #2: Processed data - aligned frame sum with dose-weighting [micrographs - single frame]) |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
NCS oper:
|
- Components
Components
| #1: Protein | Mass: 140824.406 Da / Num. of mol.: 3 / Mutation: R682G R683S R685S K986P V987P Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: S, 2 / Production host:  #2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source #3: Sugar | ChemComp-NAG / Has ligand of interest | N | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Severe acute respiratory syndrome coronavirus 2 / Type: VIRUS / Entity ID: #1 / Source: RECOMBINANT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | |||||||||||||||
| Source (natural) | Organism:  | |||||||||||||||
| Source (recombinant) | Organism:  | |||||||||||||||
| Details of virus | Empty: YES / Enveloped: YES / Isolate: OTHER / Type: VIRION | |||||||||||||||
| Buffer solution | pH: 8 | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 400 divisions/in. / Grid type: UltrAuFoil | |||||||||||||||
| Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 100 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 800 nm / C2 aperture diameter: 100 µm |
| Image recording | Average exposure time: 2.5 sec. / Electron dose: 66.5 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of real images: 1131 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 75582 | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C3 (3 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.22 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 54395 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | B value: 120 / Protocol: RIGID BODY FIT / Space: REAL / Target criteria: CC | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6VXX Pdb chain-ID: A / Accession code: 6VXX / Pdb chain residue range: 27-1147 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 120.7 Å2 | ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj






