-Search query
-Search result
Showing 1 - 50 of 205 items for (author: ni & tw)

EMDB-18135: 
Cryo-EM structure of the methanogenic Na+ translocating N5-methyl-H4MPT:CoM methyltransferase complex

PDB-8q3v: 
Cryo-EM structure of the methanogenic Na+ translocating N5-methyl-H4MPT:CoM methyltransferase complex
EMDB-43275: 
GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 1
EMDB-43276: 
GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 2

PDB-8vj6: 
GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 1

PDB-8vj7: 
GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 2

EMDB-41078: 
Acinetobacter baumannii 118362 family 2A cargo-loaded encapsulin shell
EMDB-28198: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with LLNL-199

EMDB-28199: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112

EMDB-40984: 
5TU-t1 - heterodimeric triplet polymerase ribozyme

PDB-8t2p: 
5TU-t1 - heterodimeric triplet polymerase ribozyme
EMDB-40976: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors

EMDB-40977: 
Cryo-EM structure of mink variant Y453F trimeric spike protein
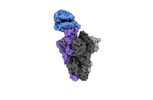
EMDB-40978: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation

EMDB-40979: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation

EMDB-40980: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation.
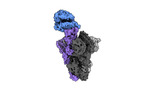
EMDB-41143: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors

PDB-8t20: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors

PDB-8t21: 
Cryo-EM structure of mink variant Y453F trimeric spike protein

PDB-8t22: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation

PDB-8t23: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation

PDB-8t25: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation.

PDB-8taz: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors

EMDB-29605: 
TpeA bound closed MthK-A88F mutant in nanodisc

EMDB-16978: 
Architecture of native chromatin fibres revealed by cryo-ET in situ

EMDB-16979: 
Architecture of native chromatin fibres revealed by cryo-ET in situ
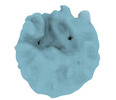
EMDB-16980: 
Architecture of native chromatin fibres revealed by cryo-ET in situ

EMDB-29813: 
mtHsp60 V72I apo

EMDB-29814: 
mtHsp60 V72I apo focus

EMDB-29815: 
ATP-bound mtHsp60 V72I

EMDB-29816: 
ATP-bound mtHsp60 V72I focus

EMDB-29817: 
ATP- and mtHsp10-bound mtHsp60 V72I

EMDB-29818: 
ATP- and mtHsp10-bound mtHsp60 V72I focus

PDB-8g7j: 
mtHsp60 V72I apo

PDB-8g7k: 
mtHsp60 V72I apo focus

PDB-8g7l: 
ATP-bound mtHsp60 V72I

PDB-8g7m: 
ATP-bound mtHsp60 V72I focus

PDB-8g7n: 
ATP- and mtHsp10-bound mtHsp60 V72I

PDB-8g7o: 
ATP- and mtHsp10-bound mtHsp60 V72I focus

EMDB-27459: 
MthK-A90L mutant in closed state with 0 Ca2+

EMDB-28776: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305

EMDB-28777: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310

EMDB-28778: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565

EMDB-28779: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296

PDB-8f0p: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305

PDB-8f0q: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310

PDB-8f0r: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565

PDB-8f0s: 
Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296

EMDB-15532: 
Plasmodium falciparum sporozoite subpellicular microtubule with interrupted luminal helix determined in situ

EMDB-15534: 
Plasmodium falciparum gametocyte subpellicular microtubule with 13 protofilaments determined in situ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
