+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ATP- and mtHsp10-bound mtHsp60 V72I focus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | chaperonin / ATPase / foldase / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcoated vesicle / isotype switching to IgG isotypes / mitochondrial unfolded protein response / TFAP2A acts as a transcriptional repressor during retinoic acid induced cell differentiation / apolipoprotein A-I binding / lipopolysaccharide receptor complex / protein import into mitochondrial intermembrane space / high-density lipoprotein particle binding / migrasome / cysteine-type endopeptidase activator activity ...coated vesicle / isotype switching to IgG isotypes / mitochondrial unfolded protein response / TFAP2A acts as a transcriptional repressor during retinoic acid induced cell differentiation / apolipoprotein A-I binding / lipopolysaccharide receptor complex / protein import into mitochondrial intermembrane space / high-density lipoprotein particle binding / migrasome / cysteine-type endopeptidase activator activity / positive regulation of T cell mediated immune response to tumor cell / Mitochondrial protein import / chaperonin ATPase / positive regulation of macrophage activation / negative regulation of execution phase of apoptosis / cellular response to interleukin-7 / MyD88-dependent toll-like receptor signaling pathway / 'de novo' protein folding / biological process involved in interaction with symbiont / sperm plasma membrane / : / apoptotic mitochondrial changes / B cell activation / B cell proliferation / positive regulation of interferon-alpha production / DNA replication origin binding / positive regulation of interleukin-10 production / apolipoprotein binding / RHOG GTPase cycle / positive regulation of execution phase of apoptosis / response to unfolded protein / Mitochondrial unfolded protein response (UPRmt) / chaperone-mediated protein complex assembly / isomerase activity / clathrin-coated pit / positive regulation of interleukin-12 production / protein folding chaperone / Mitochondrial protein degradation / intrinsic apoptotic signaling pathway / response to cold / secretory granule / T cell activation / protein maturation / sperm midpiece / ATP-dependent protein folding chaperone / lipopolysaccharide binding / positive regulation of T cell activation / positive regulation of interleukin-6 production / positive regulation of type II interferon production / osteoblast differentiation / p53 binding / unfolded protein binding / protein folding / single-stranded DNA binding / double-stranded RNA binding / protein-folding chaperone binding / protein refolding / early endosome / mitochondrial inner membrane / protein stabilization / mitochondrial matrix / ubiquitin protein ligase binding / negative regulation of apoptotic process / enzyme binding / cell surface / ATP hydrolysis activity / protein-containing complex / mitochondrion / extracellular space / RNA binding / extracellular exosome / ATP binding / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Braxton JR / Shao H / Tse E / Gestwicki JE / Southworth DR | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly. Authors: Julian R Braxton / Hao Shao / Eric Tse / Jason E Gestwicki / Daniel R Southworth /  Abstract: The mitochondrial chaperonin, mtHsp60, promotes the folding of newly imported and transiently misfolded proteins in the mitochondrial matrix, assisted by its co-chaperone mtHsp10. Despite its ...The mitochondrial chaperonin, mtHsp60, promotes the folding of newly imported and transiently misfolded proteins in the mitochondrial matrix, assisted by its co-chaperone mtHsp10. Despite its essential role in mitochondrial proteostasis, structural insights into how this chaperonin binds to clients and progresses through its ATP-dependent reaction cycle are not clear. Here, we determined cryo-electron microscopy (cryo-EM) structures of a hyperstable disease-associated mtHsp60 mutant, V72I, at three stages in this cycle. Unexpectedly, client density is identified in all states, revealing interactions with mtHsp60's apical domains and C-termini that coordinate client positioning in the folding chamber. We further identify a striking asymmetric arrangement of the apical domains in the ATP state, in which an alternating up/down configuration positions interaction surfaces for simultaneous recruitment of mtHsp10 and client retention. Client is then fully encapsulated in mtHsp60/mtHsp10, revealing prominent contacts at two discrete sites that potentially support maturation. These results identify a new role for the apical domains in coordinating client capture and progression through the cycle, and suggest a conserved mechanism of group I chaperonin function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29818.map.gz emd_29818.map.gz | 328.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29818-v30.xml emd-29818-v30.xml emd-29818.xml emd-29818.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29818_fsc.xml emd_29818_fsc.xml emd_29818_fsc_2.xml emd_29818_fsc_2.xml | 15 KB 15.1 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_29818.png emd_29818.png | 182.4 KB | ||
| Filedesc metadata |  emd-29818.cif.gz emd-29818.cif.gz | 6.7 KB | ||
| Others |  emd_29818_additional_1.map.gz emd_29818_additional_1.map.gz emd_29818_half_map_1.map.gz emd_29818_half_map_1.map.gz emd_29818_half_map_2.map.gz emd_29818_half_map_2.map.gz | 173.6 MB 322.3 MB 322.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29818 http://ftp.pdbj.org/pub/emdb/structures/EMD-29818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29818 | HTTPS FTP |
-Related structure data
| Related structure data |  8g7oMC  8g7jC  8g7kC  8g7lC  8g7mC  8g7nC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29818.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29818.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.927 Å | ||||||||||||||||||||||||||||||||||||
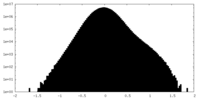
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_29818_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
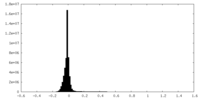
| Density Histograms |
-Half map: #2
| File | emd_29818_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
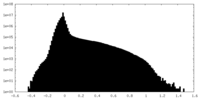
| Density Histograms |
-Half map: #1
| File | emd_29818_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
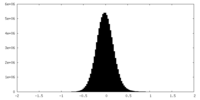
| Density Histograms |
- Sample components
Sample components
-Entire : ATP- and mtHsp10-bound mtHsp60
| Entire | Name: ATP- and mtHsp10-bound mtHsp60 |
|---|---|
| Components |
|
-Supramolecule #1: ATP- and mtHsp10-bound mtHsp60
| Supramolecule | Name: ATP- and mtHsp10-bound mtHsp60 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 60 kDa heat shock protein, mitochondrial
| Macromolecule | Name: 60 kDa heat shock protein, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO / EC number: chaperonin ATPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.204535 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AAKDVKFGAD ARALMLQGVD LLADAVAVTM GPKGRTVIIE QSWGSPKVTK DGVTVAKSID LKDKYKNIGA KLIQDVANNT NEEAGDGTT TATVLARSIA KEGFEKISKG ANPVEIRRGV MLAVDAVIAE LKKQSKPVTT PEEIAQVATI SANGDKEIGN I ISDAMKKV ...String: AAKDVKFGAD ARALMLQGVD LLADAVAVTM GPKGRTVIIE QSWGSPKVTK DGVTVAKSID LKDKYKNIGA KLIQDVANNT NEEAGDGTT TATVLARSIA KEGFEKISKG ANPVEIRRGV MLAVDAVIAE LKKQSKPVTT PEEIAQVATI SANGDKEIGN I ISDAMKKV GRKGVITVKD GKTLNDELEI IEGMKFDRGY ISPYFINTSK GQKCEFQDAY VLLSEKKISS IQSIVPALEI AN AHRKPLV IIAEDVDGEA LSTLVLNRLK VGLQVVAVKA PGFGDNRKNQ LKDMAIATGG AVFGEEGLTL NLEDVQPHDL GKV GEVIVT KDDAMLLKGK GDKAQIEKRI QEIIEQLDVT TSEYEKEKLN ERLAKLSDGV AVLKVGGTSD VEVNEKKDRV TDAL NATRA AVEEGIVLGG GCALLRCIPA LDSLTPANED QKIGIEIIKR TLKIPAMTIA KNAGVEGSLI VEKIMQSSSE VGYDA MAGD FVNMVEKGII DPTKVVRTAL LDAAGVASLL TTAEVVVTEI PKE UniProtKB: 60 kDa heat shock protein, mitochondrial |
-Macromolecule #2: 10 kDa heat shock protein, mitochondrial
| Macromolecule | Name: 10 kDa heat shock protein, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.7444 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GQAFRKFLPL FDRVLVERSA AETVTKGGIM LPEKSQGKVL QATVVAVGSG SKGKGGEIQP VSVKVGDKVL LPEYGGTKVV LDDKDYFLF RDGDILGKYV D UniProtKB: 10 kDa heat shock protein, mitochondrial |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 7 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.03 kPa / Details: Pelco easiGlow, 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: Wait time 10 sec, blot time 3 sec, blot force 0. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 2 / Number real images: 7460 / Average exposure time: 10.0 sec. / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 53937 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8g7o: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)