[English] 日本語
 Yorodumi
Yorodumi- EMDB-16979: Architecture of native chromatin fibres revealed by cryo-ET in situ -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Architecture of native chromatin fibres revealed by cryo-ET in situ | ||||||||||||
 Map data Map data | main map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | nucleosome / nucleus / histone / DNA / NUCLEAR PROTEIN | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
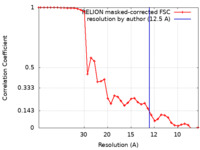
| Method | subtomogram averaging / cryo EM / Resolution: 12.5 Å | ||||||||||||
 Authors Authors | Hou Z / Zhu Y / Zhang P | ||||||||||||
| Funding support |  United Kingdom, European Union, United Kingdom, European Union,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of native chromatin fibres revealed by Cryo-ET in situ. Authors: Zhen Hou / Frank Nightingale / Yanan Zhu / Craig MacGregor-Chatwin / Peijun Zhang /  Abstract: The structure of chromatin plays pivotal roles in regulating gene transcription, DNA replication and repair, and chromosome segregation. This structure, however, remains elusive. Here, using cryo-FIB ...The structure of chromatin plays pivotal roles in regulating gene transcription, DNA replication and repair, and chromosome segregation. This structure, however, remains elusive. Here, using cryo-FIB and cryo-ET, we delineate the 3D architecture of native chromatin fibres in intact interphase human T-lymphoblasts and determine the in situ structures of nucleosomes in different conformations. These chromatin fibres are not structured as uniform 30 nm one-start or two-start filaments but are composed of relaxed, variable zigzag organizations of nucleosomes connected by straight linker DNA. Nucleosomes with little H1 and linker DNA density are distributed randomly without any spatial preference. This work will inspire future high-resolution investigations on native chromatin structures in situ at both a single-nucleosome level and a population level under many different cellular conditions in health and disease. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Structure of Native Chromatin Fibres Revealed by Cryo-ET in situ Authors: Hou Z / Nightingale F / Zhu Y / MacGregor-Chatwin C / Zhang P | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16979.map.gz emd_16979.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16979-v30.xml emd-16979-v30.xml emd-16979.xml emd-16979.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16979_fsc.xml emd_16979_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_16979.png emd_16979.png | 40.3 KB | ||
| Filedesc metadata |  emd-16979.cif.gz emd-16979.cif.gz | 3.9 KB | ||
| Others |  emd_16979_half_map_1.map.gz emd_16979_half_map_1.map.gz emd_16979_half_map_2.map.gz emd_16979_half_map_2.map.gz | 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16979 http://ftp.pdbj.org/pub/emdb/structures/EMD-16979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16979 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16979.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16979.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.36 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 2
| File | emd_16979_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
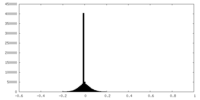
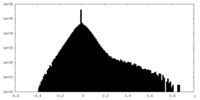
| Density Histograms |
-Half map: half map 1
| File | emd_16979_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
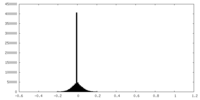
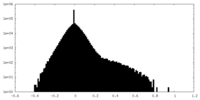
| Density Histograms |
- Sample components
Sample components
-Entire : T-Lymphoblast CEM
| Entire | Name: T-Lymphoblast CEM |
|---|---|
| Components |
|
-Supramolecule #1: T-Lymphoblast CEM
| Supramolecule | Name: T-Lymphoblast CEM / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)