-Search query
-Search result
Showing 1 - 50 of 115 items for (author: lynn & ay)

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

PDB-8sx3: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

EMDB-18342: 
E. coli DNA gyrase bound to a DNA crossover

EMDB-18565: 
E. coli DNA gyrase bound to a DNA crossover

EMDB-18566: 
Focused map of GyrA-CTD and T-segment DNA from the DNA crossover-gyrase complex

EMDB-18567: 
Focused map of GyrA-CTD from DNA crossover-gyrase complex

EMDB-18603: 
E. coli DNA gyrase bound to a linear part of a DNA minicircle

EMDB-18605: 
Asymetric subunit of E. coli DNA gyrase bound to a linear part of a DNA minicircle

PDB-8qdx: 
E. coli DNA gyrase bound to a DNA crossover

PDB-8qqs: 
E. coli DNA gyrase bound to a linear part of a DNA minicircle

PDB-8qqu: 
Asymetric subunit of E. coli DNA gyrase bound to a linear part of a DNA minicircle
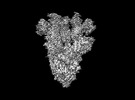
EMDB-28198: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with LLNL-199

EMDB-28199: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112

PDB-8ekd: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112

EMDB-28531: 
Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 in the 3-RBD Down conformation

EMDB-28532: 
Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 with two RBDs in the open conformation

EMDB-28533: 
Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 with two RBDs exposed

PDB-8epn: 
Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 in the 3-RBD Down conformation

PDB-8epp: 
Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 with two RBDs in the open conformation

PDB-8epq: 
Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 with two RBDs exposed

EMDB-26562: 
Cryo-EM structure of Human respiratory syncytial virus F variant (construct pXCS847A)

PDB-7uja: 
Cryo-EM structure of Human respiratory syncytial virus F variant (construct pXCS847A)

EMDB-28596: 
CryoEM Structure of NLRP3 NACHT domain in complex with G2394

PDB-8etr: 
CryoEM Structure of NLRP3 NACHT domain in complex with G2394
PDB-7n2d: 
MicroED structure of human zinc finger protein 292 segment (534-542) phased by ARCIMBOLDO-BORGES
PDB-7n2e: 
MicroED structure of human CPEB3 segment (154-161) straight polymorph
PDB-7n2f: 
MicroED structure of human CPEB3 segment (154-161) straight polymorph phased by ARCIMBOLDO-BORGES
PDB-7n2g: 
MicroED structure of human CPEB3 segment(154-161) kinked polymorph phased by ARCIMBOLDO-BORGES
PDB-7n2i: 
MicroED structure of human LECT2 (45-53) phased by ARCIMBOLDO-BORGES

PDB-7n2j: 
MicroED structure of a mutant mammalian prion segment phased by ARCIMBOLDO-BORGES
PDB-7n2k: 
MicroED structure of sequence variant of repeat segment of the yeast prion New1p phased by ARCIMBOLDO-BORGES
PDB-7n2l: 
MicroED structure of a mutant mammalian prion segment

EMDB-26180: 
Calcitonin Receptor in complex with Gs and rat amylin peptide, CT-like state

EMDB-26188: 
Calcitonin Receptor in complex with Gs and salmon calcitonin peptide

EMDB-26197: 
Human Amylin2 Receptor in complex with Gs and rat amylin peptide

PDB-7tyi: 
Calcitonin Receptor in complex with Gs and rat amylin peptide, CT-like state

PDB-7tyn: 
Calcitonin Receptor in complex with Gs and salmon calcitonin peptide

PDB-7tyx: 
Human Amylin2 Receptor in complex with Gs and rat amylin peptide

EMDB-26178: 
Human Amylin1 Receptor in complex with Gs and rat amylin peptide

EMDB-26179: 
Human Amylin2 Receptor in complex with Gs and human calcitonin peptide

EMDB-26184: 
Calcitonin Receptor in complex with Gs and rat amylin peptide, bypass motif

EMDB-26190: 
Calcitonin receptor in complex with Gs and human calcitonin peptide

EMDB-26196: 
Human Amylin1 Receptor in complex with Gs and salmon calcitonin peptide

EMDB-26199: 
Human Amylin2 Receptor in complex with Gs and salmon calcitonin peptide

EMDB-26208: 
Human Amylin3 Receptor in complex with Gs and rat amylin peptide

PDB-7tyf: 
Human Amylin1 Receptor in complex with Gs and rat amylin peptide

PDB-7tyh: 
Human Amylin2 Receptor in complex with Gs and human calcitonin peptide

PDB-7tyl: 
Calcitonin Receptor in complex with Gs and rat amylin peptide, bypass motif

PDB-7tyo: 
Calcitonin receptor in complex with Gs and human calcitonin peptide

PDB-7tyw: 
Human Amylin1 Receptor in complex with Gs and salmon calcitonin peptide
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
