-Search query
-Search result
Showing 1 - 50 of 86 items for (author: liu & js)

EMDB-43551: 
CCHFV GP38 bound with ADI-46143 and ADI-46158 Fabs

EMDB-43552: 
CCHFV GP38 bound with ADI-58062 and ADI-63530 Fabs

EMDB-43553: 
CCHFV GP38 bound with ADI-58026 and ADI-63547 Fabs

EMDB-43604: 
CCHFV GP38 bound to ADI-46152 and ADI-58048 Fabs

EMDB-41140: 
APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD

EMDB-41142: 
APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB

EMDB-16144: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1

PDB-8bon: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1

EMDB-28178: 
Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah)

EMDB-28179: 
Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41)

EMDB-28180: 
Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R)

EMDB-28181: 
Structure of lineage VII Lassa virus glycoprotein complex (strain Togo/2016/7082)

EMDB-28182: 
Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab

EMDB-28183: 
Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab

EMDB-28184: 
Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab

PDB-8ejd: 
Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah)

PDB-8eje: 
Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41)

PDB-8ejf: 
Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R)

PDB-8ejg: 
Structure of lineage VII Lassa virus glycoprotein complex (strain Togo/2016/7082)

PDB-8ejh: 
Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab

PDB-8eji: 
Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab

PDB-8ejj: 
Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab

EMDB-17154: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (consensus and constituent map 1)

EMDB-17155: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1)

EMDB-17156: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 2)

EMDB-17157: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map)

EMDB-17158: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (constituent map 2 from additional focus classification on PAS domains)

EMDB-17159: 
Cryo-EM map of MYC-MAX-OCT4-LIN28 complex

EMDB-17160: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement)

EMDB-17161: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 1)

EMDB-17162: 
MAX-MAX bound to a nucleosome at SHL+5.1 and SHL-6.9.

EMDB-17183: 
OCT4 and MYC-MAX co-bound to a nucleosome

EMDB-17184: 
MYC-MAX bound to a nucleosome at SHL+5.8

EMDB-28669: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 01)
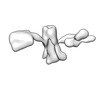
EMDB-28670: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 02)
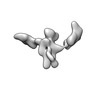
EMDB-28671: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 03)
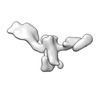
EMDB-28672: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04)

EMDB-28673: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 05)
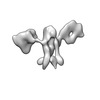
EMDB-28674: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 06)
EMDB-28675: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 07)
EMDB-28676: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 08)

EMDB-28677: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 09)

EMDB-28678: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 10)

EMDB-28679: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 11)

EMDB-28680: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 12)

EMDB-28681: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 13)

EMDB-28682: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 14)
EMDB-28683: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 15)
EMDB-28684: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 16)

EMDB-16511: 
HERV-K Gag immature lattice
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
