-Search query
-Search result
Showing all 44 items for (author: jia & xm)

EMDB-37849: 
Cryo-EM structure of CB1-beta-arrestin1 complex

EMDB-35297: 
Structural insights into human brain gut peptide cholecystokinin receptors

EMDB-33241: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex

EMDB-32769: 
Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana

EMDB-33467: 
Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana

EMDB-34023: 
EM structure of human PA28gamma

EMDB-34024: 
EM structure of human PA28gamma (wild-type)

EMDB-33359: 
Structural insights into human brain gut peptide cholecystokinin receptors

EMDB-33360: 
Structural insights into human brain gut peptide cholecystokinin receptors

EMDB-33361: 
Structural insights into human brain gut peptide cholecystokinin receptors

EMDB-32527: 
Cryo-EM structure of inactive mGlu3 bound to LY341495

EMDB-32530: 
Cryo-EM structure of LY2794193-bound mGlu3

EMDB-31241: 
Structure of the PBS-PSII supercomplex from red algae

EMDB-31242: 
Structure of the double phycobilisome-photosystem II supercomplex from red alga

EMDB-31243: 
Cryo-electron tomography of phycobilisome-photosystem II supercomplex on the thylakoid

EMDB-31244: 
Cryo-electron tomogram of phycobilisome-photosystem II supercomplex on the thylakoid

EMDB-31245: 
Cryo-electron tomogram of phycobilisome-photosystem II supercomplex on the thylakoid

EMDB-31247: 
Cryo-electron tomogram of phycobilisome-photosystem II supercomplex on the thylakoid

EMDB-30996: 
Structural insights into the activation of human calcium-sensing receptor

EMDB-30650: 
Cryo-EM structure of the Ams1 and Nbr1 complex

EMDB-30652: 
Cryo-EM structure of the Ape4 and Nbr1 complex

EMDB-30436: 
Structure of the human CLCN7-OSTM1 complex with ATP

EMDB-30438: 
Structure of the human CLCN7-OSTM1 complex with ADP

EMDB-30437: 
Structure of the human CLCN7-OSTM1 complex

EMDB-30368: 
cryo_EM map of SLC26A9

EMDB-0786: 
Cryo-EM Structure of YebT in conformation 1

EMDB-0787: 
Cryo-EM structure of YebT in conformation 2

EMDB-0788: 
Domain 5-7 of YebT in conformation 2

EMDB-9823: 
Structure of RyR2 (Ca2+ alone dataset)

EMDB-9824: 
Structure of RyR2 (F/P/Ca2+ dataset)

EMDB-9825: 
Structure of RyR2 (F/A/Ca2+ dataset)

EMDB-9826: 
Structure of RyR2 (F/C/Ca2+ dataset)

EMDB-9831: 
Structure of RyR2 (F/A/C/Ca2+ dataset)

EMDB-9833: 
Structure of RyR2 (F/apoCaM dataset)

EMDB-9834: 
Structure of RyR2 (F/A/C/L-Ca2+/apo-CaM-M dataset)

EMDB-9836: 
Structure of RyR2 (F/A/C/L-Ca2+/Ca2+CaM dataset)

EMDB-9837: 
Structure of RyR2 (F/A/C/H-Ca2+/Ca2+CaM dataset)

EMDB-9879: 
Structure of RyR2 (*F/A/C/L-Ca2+ dataset)

EMDB-9880: 
Structure of RyR2 (*F/A/C/L-Ca2+/Ca2+-CaM dataset)

EMDB-9889: 
Structure of RyR2 (P/L-Ca2+/Ca2+-CaM dataset)

EMDB-6987: 
Cryo-EM structure of a P-type ATPase
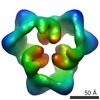
EMDB-1897: 
Reconstruction of the 3D model of AMPK trimer in basal state
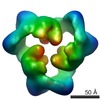
EMDB-1898: 
Reconstruction of the 3D model of AMPK trimer in ATP binding state.
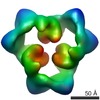
EMDB-1899: 
Reconstruction of the 3D model of AMPK trimer in AMP binding state.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
