+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32530 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of LY2794193-bound mGlu3 | |||||||||
 Map data Map data | Cryo-EM structure of LY2794193-bound mGlu3. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glutamate / G protein-coupled receptor / Cryo-EM / Selective ligand / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgroup II metabotropic glutamate receptor activity / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate receptor activity / astrocyte projection / cellular response to stress / regulation of synaptic transmission, glutamatergic / postsynaptic modulation of chemical synaptic transmission / calcium channel regulator activity ...group II metabotropic glutamate receptor activity / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate receptor activity / astrocyte projection / cellular response to stress / regulation of synaptic transmission, glutamatergic / postsynaptic modulation of chemical synaptic transmission / calcium channel regulator activity / G protein-coupled receptor activity / presynaptic membrane / scaffold protein binding / G alpha (i) signalling events / gene expression / dendritic spine / chemical synaptic transmission / postsynaptic membrane / postsynaptic density / axon / glutamatergic synapse / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.68 Å | |||||||||
 Authors Authors | Fang W / Yang F | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2022 Journal: Cell Res / Year: 2022Title: Structural basis of the activation of metabotropic glutamate receptor 3. Authors: Wei Fang / Fan Yang / Chanjuan Xu / Shenglong Ling / Li Lin / Yingxin Zhou / Wenjing Sun / Xiaomei Wang / Peng Liu / Philippe Rondard / Pan Shi / Jean-Philippe Pin / Changlin Tian / Jianfeng Liu /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32530.map.gz emd_32530.map.gz | 82.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32530-v30.xml emd-32530-v30.xml emd-32530.xml emd-32530.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32530.png emd_32530.png | 71.1 KB | ||
| Filedesc metadata |  emd-32530.cif.gz emd-32530.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32530 http://ftp.pdbj.org/pub/emdb/structures/EMD-32530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32530 | HTTPS FTP |
-Validation report
| Summary document |  emd_32530_validation.pdf.gz emd_32530_validation.pdf.gz | 426.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32530_full_validation.pdf.gz emd_32530_full_validation.pdf.gz | 426 KB | Display | |
| Data in XML |  emd_32530_validation.xml.gz emd_32530_validation.xml.gz | 6.5 KB | Display | |
| Data in CIF |  emd_32530_validation.cif.gz emd_32530_validation.cif.gz | 7.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32530 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32530 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32530 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32530 | HTTPS FTP |
-Related structure data
| Related structure data |  7wihMC  7wi6C  7wi8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32530.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32530.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of LY2794193-bound mGlu3. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM structure of mGlu3 bound with LY2794193
| Entire | Name: Cryo-EM structure of mGlu3 bound with LY2794193 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of mGlu3 bound with LY2794193
| Supramolecule | Name: Cryo-EM structure of mGlu3 bound with LY2794193 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Metabotropic glutamate receptor 3
| Macromolecule | Name: Metabotropic glutamate receptor 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100.021695 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDENLYFQG LGDHNFLRRE IKIEGDLVLG GLFPINEKGT GTEECGRINE DRGIQRLEAM LFAIDEINK DDYLLPGVKL GVHILDTCSR DTYALEQSLE FVRASLTKVD EAEYMCPDGS YAIQENIPLL IAGVIGGSYS S VSIQVANL ...String: MKTIIALSYI FCLVFADYKD DDDENLYFQG LGDHNFLRRE IKIEGDLVLG GLFPINEKGT GTEECGRINE DRGIQRLEAM LFAIDEINK DDYLLPGVKL GVHILDTCSR DTYALEQSLE FVRASLTKVD EAEYMCPDGS YAIQENIPLL IAGVIGGSYS S VSIQVANL LRLFQIPQIS YASTSAKLSD KSRYDYFART VPPDFYQAKA MAEILRFFNW TYVSTVASEG DYGETGIEAF EQ EARLRNI CIATAEKVGR SNIRKSYDSV IRELLQKPNA RVVVLFMRSD DSRELIAAAS RANASFTWVA SDGWGAQESI IKG SEHVAY GAITLELASQ PVRQFDRYFQ SLNPYNNHRN PWFRDFWEQK FQCSLQNKRN HRRVCDKHLA IDSSNYEQES KIMF VVNAV YAMAHALHKM QRTLCPNTTK LCDAMKILDG KKLYKDYLLK INFTAPFNPN KDADSIVKFD TFGDGMGRYN VFNFQ NVGG KYSYLKVGHW AETLSLDVNS IHWSRNSVPT SQCSDPCAPN EMKNMQPGDV CCWICIPCEP YEYLADEFTC MDCGSG QWP TADLTGCYDL PEDYIRWEDA WAIGPVTIAC LGFMCTCMVV TVFIKHNNTP LVKASGRELC YILLFGVGLS YCMTFFF IA KPSPVICALR RLGLGSSFAI CYSALLTKTN CIARIFDGVK NGAQRPKFIS PSSQVFICLG LILVQIVMVS VWLILEAP G TRRYTLAEKR ETVILKCNVK DSSMLISLTY DVILVILCTV YAFKTRKCPE NFNEAKFIGF TMYTTCIIWL AFLPIFYVT SSDYRVQTTT MCISVSLSGF VVLGCLFAPK VHIILFQPQK NVVTHRLHLN RFSVSGTGTT YSQSSASTYV PTVCNGREVL DSTTSSL UniProtKB: Metabotropic glutamate receptor 3 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bi...
| Macromolecule | Name: (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid type: ligand / ID: 3 / Number of copies: 2 / Formula: CWY |
|---|---|
| Molecular weight | Theoretical: 334.324 Da |
| Chemical component information | 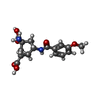 ChemComp-CWY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 57.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.68 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 288762 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















