[English] 日本語
 Yorodumi
Yorodumi- EMDB-30996: Structural insights into the activation of human calcium-sensing ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30996 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural insights into the activation of human calcium-sensing receptor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G-protein-coupled receptor (GPCR) / Calcium-sensing receptor (CaSR) / cryo-electron microscopy (cryo-EM) / calcium ions / nanobody / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of presynaptic membrane potential / bile acid secretion / chemosensory behavior / response to fibroblast growth factor / cellular response to peptide / cellular response to vitamin D / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / calcium ion import / positive regulation of positive chemotaxis ...regulation of presynaptic membrane potential / bile acid secretion / chemosensory behavior / response to fibroblast growth factor / cellular response to peptide / cellular response to vitamin D / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / calcium ion import / positive regulation of positive chemotaxis / fat pad development / cellular response to hepatocyte growth factor stimulus / amino acid binding / branching morphogenesis of an epithelial tube / positive regulation of calcium ion import / positive regulation of vasoconstriction / regulation of calcium ion transport / cellular response to low-density lipoprotein particle stimulus / anatomical structure morphogenesis / detection of calcium ion / JNK cascade / axon terminus / ossification / chloride transmembrane transport / response to ischemia / cellular response to glucose stimulus / positive regulation of insulin secretion / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / integrin binding / vasodilation / intracellular calcium ion homeostasis / presynaptic membrane / G alpha (i) signalling events / cellular response to hypoxia / phospholipase C-activating G protein-coupled receptor signaling pathway / basolateral plasma membrane / G alpha (q) signalling events / transmembrane transporter binding / positive regulation of ERK1 and ERK2 cascade / apical plasma membrane / G protein-coupled receptor signaling pathway / neuronal cell body / positive regulation of cell population proliferation / calcium ion binding / positive regulation of gene expression / protein kinase binding / glutamatergic synapse / cell surface / protein homodimerization activity / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Geng Y / Chen XC | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Structural insights into the activation of human calcium-sensing receptor. Authors: Xiaochen Chen / Lu Wang / Qianqian Cui / Zhanyu Ding / Li Han / Yongjun Kou / Wenqing Zhang / Haonan Wang / Xiaomin Jia / Mei Dai / Zhenzhong Shi / Yuying Li / Xiyang Li / Yong Geng /  Abstract: Human calcium-sensing receptor (CaSR) is a G-protein-coupled receptor that maintains Ca homeostasis in serum. Here, we present the cryo-electron microscopy structures of the CaSR in the inactive and ...Human calcium-sensing receptor (CaSR) is a G-protein-coupled receptor that maintains Ca homeostasis in serum. Here, we present the cryo-electron microscopy structures of the CaSR in the inactive and agonist+PAM bound states. Complemented with previously reported structures of CaSR, we show that in addition to the full inactive and active states, there are multiple intermediate states during the activation of CaSR. We used a negative allosteric nanobody to stabilize the CaSR in the fully inactive state and found a new binding site for Ca ion that acts as a composite agonist with L-amino acid to stabilize the closure of active Venus flytraps. Our data show that agonist binding leads to compaction of the dimer, proximity of the cysteine-rich domains, large-scale transitions of seven-transmembrane domains, and inter- and intrasubunit conformational changes of seven-transmembrane domains to accommodate downstream transducers. Our results reveal the structural basis for activation mechanisms of CaSR and clarify the mode of action of Ca ions and L-amino acid leading to the activation of the receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30996.map.gz emd_30996.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30996-v30.xml emd-30996-v30.xml emd-30996.xml emd-30996.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30996.png emd_30996.png | 43.5 KB | ||
| Filedesc metadata |  emd-30996.cif.gz emd-30996.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30996 http://ftp.pdbj.org/pub/emdb/structures/EMD-30996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30996 | HTTPS FTP |
-Validation report
| Summary document |  emd_30996_validation.pdf.gz emd_30996_validation.pdf.gz | 499.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30996_full_validation.pdf.gz emd_30996_full_validation.pdf.gz | 498.8 KB | Display | |
| Data in XML |  emd_30996_validation.xml.gz emd_30996_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  emd_30996_validation.cif.gz emd_30996_validation.cif.gz | 7.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30996 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30996 | HTTPS FTP |
-Related structure data
| Related structure data |  7e6tMC  7e6uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30996.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30996.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.105 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CaSR, TNCA, Ca, PO4
| Entire | Name: CaSR, TNCA, Ca, PO4 |
|---|---|
| Components |
|
-Supramolecule #1: CaSR, TNCA, Ca, PO4
| Supramolecule | Name: CaSR, TNCA, Ca, PO4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Extracellular calcium-sensing receptor
| Macromolecule | Name: Extracellular calcium-sensing receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 97.326094 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YGPDQRAQKK GDIILGGLFP IHFGVAAKDQ DLKSRPESVE CIRYNFRGFR WLQAMIFAIE EINSSPALLP NLTLGYRIFD TCNTVSKAL EATLSFVAQN KIDSLNLDEF CNCSEHIPST IAVVGATGSG VSTAVANLLG LFYIPQVSYA SSSRLLSNKN Q FKSFLRTI ...String: YGPDQRAQKK GDIILGGLFP IHFGVAAKDQ DLKSRPESVE CIRYNFRGFR WLQAMIFAIE EINSSPALLP NLTLGYRIFD TCNTVSKAL EATLSFVAQN KIDSLNLDEF CNCSEHIPST IAVVGATGSG VSTAVANLLG LFYIPQVSYA SSSRLLSNKN Q FKSFLRTI PNDEHQATAM ADIIEYFRWN WVGTIAADDD YGRPGIEKFR EEAEERDICI DFSELISQYS DEEEIQHVVE VI QNSTAKV IVVFSSGPDL EPLIKEIVRR NITGKIWLAS EAWASSSLIA MPQYFHVVGG TIGFALKAGQ IPGFREFLKK VHP RKSVHN GFAKEFWEET FNCHLQEGAK GPLPVDTFLR GHEESGDRFS NSSTAFRPLC TGDENISSVE TPYIDYTHLR ISYN VYLAV YSIAHALQDI YTCLPGRGLF TNGSCADIKK VEAWQVLKHL RHLNFTNNMG EQVTFDECGD LVGNYSIINW HLSPE DGSI VFKEVGYYNV YAKKGERLFI NEEKILWSGF SREVPFSNCS RDCLAGTRKG IIEGEPTCCF ECVECPDGEY SDETDA SAC NKCPDDFWSN ENHTSCIAKE IEFLSWTEPF GIALTLFAVL GIFLTAFVLG VFIKFRNTPI VKATNRELSY LLLFSLL CC FSSSLFFIGE PQDWTCRLRQ PAFGISFVLC ISCILVKTNR VLLVFEAKIP TSFHRKWWGL NLQFLLVFLC TFMQIVIC V IWLYTAPPSS YRNQELEDEI IFITCHEGSL MALGFLIGYT CLLAAICFFF AFKSRKLPEN FNEAKFITFS MLIFFIVWI SFIPAYASTY GKFVSAVEVI AILAASFGLL ACIFFNKIYI ILFKPSRNTI EAAADYKDDD DK UniProtKB: Extracellular calcium-sensing receptor |
-Macromolecule #2: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: CYCLOMETHYLTRYPTOPHAN
| Macromolecule | Name: CYCLOMETHYLTRYPTOPHAN / type: ligand / ID: 5 / Number of copies: 2 / Formula: TCR |
|---|---|
| Molecular weight | Theoretical: 216.236 Da |
| Chemical component information | 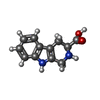 ChemComp-TCR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: ZERNIKE PHASE PLATE |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















