[English] 日本語
 Yorodumi
Yorodumi- EMDB-8954: Handover mechanism of the growing pilus by the bacterial outer me... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8954 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
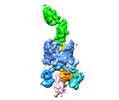
| Title | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pili / chaperone / usher / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationfimbrial usher porin activity / pilus assembly / pilus tip / mechanosensory behavior / cell adhesion involved in single-species biofilm formation / pilus / : / cell-substrate adhesion / D-mannose binding / host cell membrane ...fimbrial usher porin activity / pilus assembly / pilus tip / mechanosensory behavior / cell adhesion involved in single-species biofilm formation / pilus / : / cell-substrate adhesion / D-mannose binding / host cell membrane / protein folding chaperone / cell outer membrane / cell wall organization / outer membrane-bounded periplasmic space / cell adhesion Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.1 Å | |||||||||
 Authors Authors | Du M / Yuan Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD. Authors: Minge Du / Zuanning Yuan / Hongjun Yu / Nadine Henderson / Samema Sarowar / Gongpu Zhao / Glenn T Werneburg / David G Thanassi / Huilin Li /  Abstract: Pathogenic bacteria such as Escherichia coli assemble surface structures termed pili, or fimbriae, to mediate binding to host-cell receptors. Type 1 pili are assembled via the conserved chaperone- ...Pathogenic bacteria such as Escherichia coli assemble surface structures termed pili, or fimbriae, to mediate binding to host-cell receptors. Type 1 pili are assembled via the conserved chaperone-usher pathway. The outer-membrane usher FimD recruits pilus subunits bound by the chaperone FimC via the periplasmic N-terminal domain of the usher. Subunit translocation through the β-barrel channel of the usher occurs at the two C-terminal domains (which we label CTD1 and CTD2) of this protein. How the chaperone-subunit complex bound to the N-terminal domain is handed over to the C-terminal domains, as well as the timing of subunit polymerization into the growing pilus, have previously been unclear. Here we use cryo-electron microscopy to capture a pilus assembly intermediate (FimD-FimC-FimF-FimG-FimH) in a conformation in which FimD is in the process of handing over the chaperone-bound end of the growing pilus to the C-terminal domains. In this structure, FimF has already polymerized with FimG, and the N-terminal domain of FimD swings over to bind CTD2; the N-terminal domain maintains contact with FimC-FimF, while at the same time permitting access to the C-terminal domains. FimD has an intrinsically disordered N-terminal tail that precedes the N-terminal domain. This N-terminal tail folds into a helical motif upon recruiting the FimC-subunit complex, but reorganizes into a loop to bind CTD2 during handover. Because both the N-terminal and C-terminal domains of FimD are bound to the end of the growing pilus, the structure further suggests a mechanism for stabilizing the assembly intermediate to prevent the pilus fibre diffusing away during the incorporation of thousands of subunits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8954.map.gz emd_8954.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8954-v30.xml emd-8954-v30.xml emd-8954.xml emd-8954.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8954.png emd_8954.png | 103.7 KB | ||
| Filedesc metadata |  emd-8954.cif.gz emd-8954.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8954 http://ftp.pdbj.org/pub/emdb/structures/EMD-8954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8954 | HTTPS FTP |
-Related structure data
| Related structure data |  6e15MC  8953C  6e14C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8954.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8954.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.074 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : FimDCFGH tip complex
| Entire | Name: FimDCFGH tip complex |
|---|---|
| Components |
|
-Supramolecule #1: FimDCFGH tip complex
| Supramolecule | Name: FimDCFGH tip complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Chaperone protein FimC
| Macromolecule | Name: Chaperone protein FimC / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.716869 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNKNVNVRK SQEITFCLLA GILMFMAMMV AGRAEAGVAL GATRVIYPAG QKQEQLAVTN NDENSTYLIQ SWVENADGVK DGRFIVTPP LFAMKGKKEN TLRILDATNN QLPQDRESLF WMNVKAIPSM DKSKLTENTL QLAIISRIKL YYRPAKLALP P DQAAEKLR ...String: MSNKNVNVRK SQEITFCLLA GILMFMAMMV AGRAEAGVAL GATRVIYPAG QKQEQLAVTN NDENSTYLIQ SWVENADGVK DGRFIVTPP LFAMKGKKEN TLRILDATNN QLPQDRESLF WMNVKAIPSM DKSKLTENTL QLAIISRIKL YYRPAKLALP P DQAAEKLR FRRSANSLTL INPTPYYLTV TELNAGTRVL ENALVPPMGE STVKLPSDAG SNITYRTIND YGALTPKMTG VM E UniProtKB: Chaperone protein FimC |
-Macromolecule #2: Fimbrial biogenesis outer membrane usher protein
| Macromolecule | Name: Fimbrial biogenesis outer membrane usher protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 96.705109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYLNLRLYQ RNTQCLHIRK HRLAGFFVRL FVACAFAAQA SLSSAELYFN PRFLADDPQA VADLSRFENG QELPPGTYRV DIYLNNGYM ATRDVTFNTG DSEQGIVPCL TRAQLASMGL NTASVAGMNL LADDACVPLT TMVQDATAHL DVGQQRLNLT I PQAFMSNR ...String: MSYLNLRLYQ RNTQCLHIRK HRLAGFFVRL FVACAFAAQA SLSSAELYFN PRFLADDPQA VADLSRFENG QELPPGTYRV DIYLNNGYM ATRDVTFNTG DSEQGIVPCL TRAQLASMGL NTASVAGMNL LADDACVPLT TMVQDATAHL DVGQQRLNLT I PQAFMSNR ARGYIPPELW DPGINAGLLN YNFSGNSVQN RIGGNSHYAY LNLQSGLNIG AWRLRDNTTW SYNSSDRSSG SK NKWQHIN TWLERDIIPL RSRLTLGDGY TQGDIFDGIN FRGAQLASDD NMLPDSQRGF APVIHGIARG TAQVTIKQNG YDI YNSTVP PGPFTINDIY AAGNSGDLQV TIKEADGSTQ IFTVPYSSVP LLQREGHTRY SITAGEYRSG NAQQEKPRFF QSTL LHGLP AGWTIYGGTQ LADRYRAFNF GIGKNMGALG ALSVDMTQAN STLPDDSQHD GQSVRFLYNK SLNESGTNIQ LVGYR YSTS GYFNFADTTY SRMNGYNIET QDGVIQVKPK FTDYYNLAYN KRGKLQLTVT QQLGRTSTLY LSGSHQTYWG TSNVDE QFQ AGLNTAFEDI NWTLSYSLTK NAWQKGRDQM LALNVNIPFS HWLRSDSKSQ WRHASASYSM SHDLNGRMTN LAGVYGT LL EDNNLSYSVQ TGYAGGGDGN SGSTGYATLN YRGGYGNANI GYSHSDDIKQ LYYGVSGGVL AHANGVTLGQ PLNDTVVL V KAPGAKDAKV ENQTGVRTDW RGYAVLPYAT EYRENRVALD TNTLADNVDL DNAVANVVPT RGAIVRAEFK ARVGIKLLM TLTHNNKPLP FGAMVTSESS QSSGIVADNG QVYLSGMPLA GKVQVKWGEE ENAHCVANYQ LPPESQQQLL TQLSAECRS UniProtKB: Fimbrial biogenesis outer membrane usher protein |
-Macromolecule #3: Protein FimF
| Macromolecule | Name: Protein FimF / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.379371 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAADSTITIR GYVRDNGCSV AAESTNFTVD LMENAAKQFN NIGATTPVVP FRILLSPCGN AVSAVKVGFT GVADSHNANL LALENTVSA ASGLGIQLLN EQQNQIPLNA PSSALSWTTL TPGKPNTLNF YARLMATQVP VTAGHINATA TFTLEYQ UniProtKB: Protein FimF |
-Macromolecule #4: Protein FimG
| Macromolecule | Name: Protein FimG / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.190834 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAILALASA TIQAADVTIT VNGKVVAKPC TVSTTNATVD LGDLYSFSLM SAGAASAWHD VALELTNCPV GTSRVTASFS GAADSTGYY KNQGTAQNIQ LELQDDSGNT LNTGATKTVQ VDDSSQSAHF PLQVRALTVN GGATQGTIQA VISITYTYS UniProtKB: Protein FimG |
-Macromolecule #5: Type 1 fimbrin D-mannose specific adhesin
| Macromolecule | Name: Type 1 fimbrin D-mannose specific adhesin / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.48826 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKRVITLFAV LLMGWSVNAW SFACKTANGT AIPIGGGSAN VYVNLAPVVN VGQNLVVDLS TQIFCHNDYP ETITDYVTLQ RGSAYGGVL SNFSGTVKYS GSSYPFPTTS ETPRVVYNSR TDKPWPVALY LTPVSSAGGV AIKAGSLIAV LILRQTNNYN S DDFQFVWN ...String: MKRVITLFAV LLMGWSVNAW SFACKTANGT AIPIGGGSAN VYVNLAPVVN VGQNLVVDLS TQIFCHNDYP ETITDYVTLQ RGSAYGGVL SNFSGTVKYS GSSYPFPTTS ETPRVVYNSR TDKPWPVALY LTPVSSAGGV AIKAGSLIAV LILRQTNNYN S DDFQFVWN IYANNDVVVP TGGCDVSARD VTVTLPDYPG SVPIPLTVYC AKSQNLGYYL SGTTADAGNS IFTNTASFSP AQ GVGVQLT RNGTIIPANN TVSLGAVGTS AVSLGLTANY ARTGGQVTAG NVQSIIGVTF VYQ UniProtKB: Type 1 fimbrin D-mannose specific adhesin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















