+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CB1-beta-arrestin1 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cannabinoid receptor / arrestin / biased signal / class A GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTGFBR3 regulates TGF-beta signaling / MAP2K and MAPK activation / Activation of SMO / cannabinoid signaling pathway / Golgi Associated Vesicle Biogenesis / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / Lysosome Vesicle Biogenesis / retrograde trans-synaptic signaling by endocannabinoid / Vasopressin-like receptors ...TGFBR3 regulates TGF-beta signaling / MAP2K and MAPK activation / Activation of SMO / cannabinoid signaling pathway / Golgi Associated Vesicle Biogenesis / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / Lysosome Vesicle Biogenesis / retrograde trans-synaptic signaling by endocannabinoid / Vasopressin-like receptors / regulation of systemic arterial blood pressure by vasopressin / vasopressin receptor activity / cannabinoid receptor activity / AP-2 adaptor complex binding / Ub-specific processing proteases / clathrin coat of coated pit / clathrin heavy chain binding / Cargo recognition for clathrin-mediated endocytosis / hemostasis / desensitization of G protein-coupled receptor signaling pathway / regulation of presynaptic cytosolic calcium ion concentration / telencephalon development / Clathrin-mediated endocytosis / clathrin-dependent endocytosis / acetylcholine receptor binding / Class A/1 (Rhodopsin-like receptors) / axonal fasciculation / G protein-coupled receptor internalization / inositol hexakisphosphate binding / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (s) signalling events / clathrin binding / small molecule binding / pseudopodium / phosphatidylinositol-3,4,5-trisphosphate binding / positive regulation of systemic arterial blood pressure / positive regulation of intracellular signal transduction / positive regulation of receptor internalization / positive regulation of vasoconstriction / negative regulation of Notch signaling pathway / endocytic vesicle / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / activation of adenylate cyclase activity / cellular response to hormone stimulus / response to cytokine / clathrin-coated endocytic vesicle membrane / G protein-coupled receptor binding / receptor internalization / G protein-coupled receptor activity / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / positive regulation of protein phosphorylation / Vasopressin regulates renal water homeostasis via Aquaporins / Cargo recognition for clathrin-mediated endocytosis / actin cytoskeleton / glucose homeostasis / protein transport / Clathrin-mediated endocytosis / growth cone / presynaptic membrane / cytoplasmic vesicle / G alpha (i) signalling events / G alpha (s) signalling events / ubiquitin-dependent protein catabolic process / molecular adaptor activity / mitochondrial outer membrane / positive regulation of ERK1 and ERK2 cascade / endosome / membrane raft / G protein-coupled receptor signaling pathway / negative regulation of cell population proliferation / positive regulation of cell population proliferation / positive regulation of gene expression / perinuclear region of cytoplasm / glutamatergic synapse / endoplasmic reticulum / Golgi apparatus / signal transduction / identical protein binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Liao Y / Zhang H / Shen Q / Cai C | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Snapshot of the cannabinoid receptor 1-arrestin complex unravels the biased signaling mechanism. Authors: Yu-Ying Liao / Huibing Zhang / Qingya Shen / Chenxi Cai / Yu Ding / Dan-Dan Shen / Jia Guo / Jiao Qin / Yingjun Dong / Yan Zhang / Xiao-Ming Li /  Abstract: Cannabis activates the cannabinoid receptor 1 (CB1), which elicits analgesic and emotion regulation benefits, along with adverse effects, via G and β-arrestin signaling pathways. However, the lack ...Cannabis activates the cannabinoid receptor 1 (CB1), which elicits analgesic and emotion regulation benefits, along with adverse effects, via G and β-arrestin signaling pathways. However, the lack of understanding of the mechanism of β-arrestin-1 (βarr1) coupling and signaling bias has hindered drug development targeting CB1. Here, we present the high-resolution cryo-electron microscopy structure of CB1-βarr1 complex bound to the synthetic cannabinoid MDMB-Fubinaca (FUB), revealing notable differences in the transducer pocket and ligand-binding site compared with the G protein complex. βarr1 occupies a wider transducer pocket promoting substantial outward movement of the TM6 and distinctive twin toggle switch rearrangements, whereas FUB adopts a different pose, inserting more deeply than the G-coupled state, suggesting the allosteric correlation between the orthosteric binding pocket and the partner protein site. Taken together, our findings unravel the molecular mechanism of signaling bias toward CB1, facilitating the development of CB1 agonists. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37849.map.gz emd_37849.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37849-v30.xml emd-37849-v30.xml emd-37849.xml emd-37849.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37849.png emd_37849.png | 55.6 KB | ||
| Filedesc metadata |  emd-37849.cif.gz emd-37849.cif.gz | 7.2 KB | ||
| Others |  emd_37849_half_map_1.map.gz emd_37849_half_map_1.map.gz emd_37849_half_map_2.map.gz emd_37849_half_map_2.map.gz | 39.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37849 http://ftp.pdbj.org/pub/emdb/structures/EMD-37849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37849 | HTTPS FTP |
-Related structure data
| Related structure data |  8wu1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37849.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37849.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37849_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
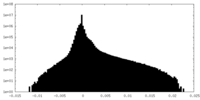
| Density Histograms |
-Half map: #1
| File | emd_37849_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of human cannabinoid receptor 1 with effector p...
| Entire | Name: Cryo-EM structure of human cannabinoid receptor 1 with effector protein |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of human cannabinoid receptor 1 with effector p...
| Supramolecule | Name: Cryo-EM structure of human cannabinoid receptor 1 with effector protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: MDMB-Fubinaca, cannabinoid receptor 1, arrestin2, Fab30 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Beta-arrestin-1
| Macromolecule | Name: Beta-arrestin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.135273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGDKGTRVFK KASPNGKLTV YLGKRDFVDH IDLVEPVDGV VLVDPEYLKE RRVYVTLTCA FRYGREDLDV LGLTFRKDLF VANVQSFPP APEDKKPLTR LQERLIKKLG EHAYPFTFEI PPNLPCSVTL QPGPEDTGKA CGVDYEVKAF CAENLEEKIH K RNSVRLVI ...String: MGDKGTRVFK KASPNGKLTV YLGKRDFVDH IDLVEPVDGV VLVDPEYLKE RRVYVTLTCA FRYGREDLDV LGLTFRKDLF VANVQSFPP APEDKKPLTR LQERLIKKLG EHAYPFTFEI PPNLPCSVTL QPGPEDTGKA CGVDYEVKAF CAENLEEKIH K RNSVRLVI EKVQYAPERP GPQPTAETTR QFLMSDKPLH LEASLDKEIY YHGEPISVNV HVTNNTNKTV KKIKISVRQY AD ICLFNTA QYKCPVAMEE ADDTVAPSST FCKVYTLTPF LANNREKRGL ALDGKLKHED TNLASSTLLR EGANREILGI IVS YKVKVK LVVSRGGLLG DLASSDVAVE LPFTLMHPKP KEEPPHREVP EHETPVDTNL IELDTNDDDA AAEDFAR UniProtKB: Beta-arrestin-1 |
-Macromolecule #2: Fab30 heavy chain
| Macromolecule | Name: Fab30 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.650502 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYYGY TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSRQFWYSG LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYYGY TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSRQFWYSG LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDK THHHHHHHHH |
-Macromolecule #3: Fab30 light chain
| Macromolecule | Name: Fab30 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.435064 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYKYVPVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYKYVPVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: Cannabinoid receptor 1,Vasopressin V2 receptor
| Macromolecule | Name: Cannabinoid receptor 1,Vasopressin V2 receptor / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.310129 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSILDGLAD TTFRTITTDL LYVGSNDIQY EDIKGDMASK LGYFPQKFPL TSFRGSPFQE KMTAGDNPQL VPADQVNITE FYNKSLSSF KENEENIQCG ENFMDIECFM VLNPSQQLAI AVLSLTLGTF TVLENLLVLC VILHSRSLRC RPSYHFIGSL A VADLLGSV ...String: MKSILDGLAD TTFRTITTDL LYVGSNDIQY EDIKGDMASK LGYFPQKFPL TSFRGSPFQE KMTAGDNPQL VPADQVNITE FYNKSLSSF KENEENIQCG ENFMDIECFM VLNPSQQLAI AVLSLTLGTF TVLENLLVLC VILHSRSLRC RPSYHFIGSL A VADLLGSV IFVYSFIDFH VFHRKDSRNV FLFKLGGVTA SFTASVGSLF LTAIDRYISI HRPLAYKRIV TRPKAVVAFC LM WTIAIVI AVLPLLGWNC EKLQSVCSDI FPHIDETYLM FWIGVTSVLL LFIVYAYMYI LWKAHSHAVR MIQRGTQKSI IIH TSEDGK VQVTRPDQAR MDIRLAKTLV LILVVLIICW GPLLAIMVYD VFGKMNKLIK TVFAFCSMLC LLNSTVNPII YALR SKDLR HAFRSMFPCA RGRTPPSLGP QDE(SEP)C(TPO)(TPO)A(SEP)(SEP) (SEP)LAKDTSS UniProtKB: Cannabinoid receptor 1, Vasopressin V2 receptor |
-Macromolecule #5: methyl N-{1-[(4-fluorophenyl)methyl]-1H-indazole-3-carbonyl}-3-me...
| Macromolecule | Name: methyl N-{1-[(4-fluorophenyl)methyl]-1H-indazole-3-carbonyl}-3-methyl-L-valinate type: ligand / ID: 5 / Number of copies: 1 / Formula: KCA |
|---|---|
| Molecular weight | Theoretical: 397.443 Da |
| Chemical component information | 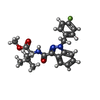 ChemComp-KCA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)