+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8wu1 | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CB1-beta-arrestin1 complex | |||||||||||||||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / cannabinoid receptor / arrestin / biased signal / class A GPCR | |||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTGFBR3 regulates TGF-beta signaling / MAP2K and MAPK activation / Activation of SMO / cannabinoid signaling pathway / Golgi Associated Vesicle Biogenesis / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / Lysosome Vesicle Biogenesis / Vasopressin-like receptors / regulation of systemic arterial blood pressure by vasopressin ...TGFBR3 regulates TGF-beta signaling / MAP2K and MAPK activation / Activation of SMO / cannabinoid signaling pathway / Golgi Associated Vesicle Biogenesis / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / Lysosome Vesicle Biogenesis / Vasopressin-like receptors / regulation of systemic arterial blood pressure by vasopressin / vasopressin receptor activity / retrograde trans-synaptic signaling by endocannabinoid / cannabinoid receptor activity / AP-2 adaptor complex binding / Ub-specific processing proteases / clathrin coat of coated pit / clathrin heavy chain binding / Cargo recognition for clathrin-mediated endocytosis / hemostasis / desensitization of G protein-coupled receptor signaling pathway / telencephalon development / regulation of presynaptic cytosolic calcium ion concentration / Clathrin-mediated endocytosis / clathrin-dependent endocytosis / acetylcholine receptor binding / Class A/1 (Rhodopsin-like receptors) / G protein-coupled receptor internalization / axonal fasciculation / inositol hexakisphosphate binding / regulation of metabolic process / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (s) signalling events / clathrin binding / small molecule binding / positive regulation of vasoconstriction / pseudopodium / phosphatidylinositol-3,4,5-trisphosphate binding / positive regulation of systemic arterial blood pressure / positive regulation of intracellular signal transduction / positive regulation of receptor internalization / negative regulation of Notch signaling pathway / endocytic vesicle / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / activation of adenylate cyclase activity / cellular response to hormone stimulus / response to cytokine / clathrin-coated endocytic vesicle membrane / G protein-coupled receptor binding / G protein-coupled receptor activity / receptor internalization / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / positive regulation of protein phosphorylation / Vasopressin regulates renal water homeostasis via Aquaporins / Cargo recognition for clathrin-mediated endocytosis / actin cytoskeleton / glucose homeostasis / protein transport / Clathrin-mediated endocytosis / growth cone / presynaptic membrane / cytoplasmic vesicle / ubiquitin-dependent protein catabolic process / G alpha (i) signalling events / G alpha (s) signalling events / molecular adaptor activity / mitochondrial outer membrane / positive regulation of ERK1 and ERK2 cascade / endosome / membrane raft / G protein-coupled receptor signaling pathway / negative regulation of cell population proliferation / positive regulation of cell population proliferation / positive regulation of gene expression / perinuclear region of cytoplasm / glutamatergic synapse / endoplasmic reticulum / Golgi apparatus / signal transduction / identical protein binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Liao, Y. / Zhang, H. / Shen, Q. / Cai, C. | |||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  China, 1items China, 1items
| |||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Snapshot of the cannabinoid receptor 1-arrestin complex unravels the biased signaling mechanism. Authors: Yu-Ying Liao / Huibing Zhang / Qingya Shen / Chenxi Cai / Yu Ding / Dan-Dan Shen / Jia Guo / Jiao Qin / Yingjun Dong / Yan Zhang / Xiao-Ming Li /  Abstract: Cannabis activates the cannabinoid receptor 1 (CB1), which elicits analgesic and emotion regulation benefits, along with adverse effects, via G and β-arrestin signaling pathways. However, the lack ...Cannabis activates the cannabinoid receptor 1 (CB1), which elicits analgesic and emotion regulation benefits, along with adverse effects, via G and β-arrestin signaling pathways. However, the lack of understanding of the mechanism of β-arrestin-1 (βarr1) coupling and signaling bias has hindered drug development targeting CB1. Here, we present the high-resolution cryo-electron microscopy structure of CB1-βarr1 complex bound to the synthetic cannabinoid MDMB-Fubinaca (FUB), revealing notable differences in the transducer pocket and ligand-binding site compared with the G protein complex. βarr1 occupies a wider transducer pocket promoting substantial outward movement of the TM6 and distinctive twin toggle switch rearrangements, whereas FUB adopts a different pose, inserting more deeply than the G-coupled state, suggesting the allosteric correlation between the orthosteric binding pocket and the partner protein site. Taken together, our findings unravel the molecular mechanism of signaling bias toward CB1, facilitating the development of CB1 agonists. | |||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8wu1.cif.gz 8wu1.cif.gz | 186.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8wu1.ent.gz pdb8wu1.ent.gz | 137.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8wu1.json.gz 8wu1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wu/8wu1 https://data.pdbj.org/pub/pdb/validation_reports/wu/8wu1 ftp://data.pdbj.org/pub/pdb/validation_reports/wu/8wu1 ftp://data.pdbj.org/pub/pdb/validation_reports/wu/8wu1 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  37849MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 44135.273 Da / Num. of mol.: 1 / Mutation: R169E, I386A, V387A, F388A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Antibody | Mass: 25650.502 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  |
| #3: Antibody | Mass: 23435.064 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  |
| #4: Protein | Mass: 50310.129 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CNR1, AVPR2 / Production host: Homo sapiens (human) / Gene: CNR1, AVPR2 / Production host:  |
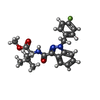
| #5: Chemical | ChemComp-KCA / |
| Has ligand of interest | N |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-EM structure of human cannabinoid receptor 1 with effector protein Type: COMPLEX Details: MDMB-Fubinaca, cannabinoid receptor 1, arrestin2, Fab30 Entity ID: #1-#4 / Source: RECOMBINANT | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| ||||||||||||
| Source (recombinant) |
| ||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||
| Specimen | Conc.: 5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 5000 nm / Nominal defocus min: 1200 nm |
| Image recording | Electron dose: 52 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 110196 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj