-Search query
-Search result
Showing 1 - 50 of 3,649 items for (author: huang & m)

EMDB-44482: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab

EMDB-44484: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs

EMDB-44491: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

PDB-9ber: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab

PDB-9bew: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs

PDB-9bf6: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

EMDB-39645: 
The structure of HKU1-B S protein with bsAb1

EMDB-39646: 
the complex structure of the H4B6 Fab with the RBD of Omicron BA.5 S protein

PDB-8yww: 
The structure of HKU1-B S protein with bsAb1

PDB-8ywx: 
the complex structure of the H4B6 Fab with the RBD of Omicron BA.5 S protein

EMDB-43139: 
SARS-CoV-2 Spike S2 bound to Fab 54043-5

EMDB-32979: 
Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10)

PDB-7x35: 
Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10)

EMDB-44635: 
Inactive mu opioid receptor bound to Nb6, naloxone and NAM

PDB-9bjk: 
Inactive mu opioid receptor bound to Nb6, naloxone and NAM

EMDB-38559: 
Structure of the sea urchin spSLC9C1 in state-2 w/o cAMP dimer

EMDB-38565: 
Structure of the sea urchin spSLC9C1 in state-2 w/o cAMP protomer

EMDB-38568: 
Structure of the sea urchin spSLC9C1 in state-1 w/ cAMP dimer

EMDB-38569: 
Structure of the sea urchin spSLC9C1 in state-1 w/ cAMP protomer

EMDB-38570: 
Structure of the sea urchin spSLC9C1 in state-2 w/ cAMP dimer

EMDB-38571: 
Structure of the sea urchin spSLC9C1 in state-3 w/ cAMP dimer

PDB-8xpq: 
Structure of the sea urchin spSLC9C1 in state-2 w/o cAMP dimer

PDB-8xq4: 
Structure of the sea urchin spSLC9C1 in state-2 w/o cAMP protomer

PDB-8xq7: 
Structure of the sea urchin spSLC9C1 in state-1 w/ cAMP dimer

PDB-8xq8: 
Structure of the sea urchin spSLC9C1 in state-1 w/ cAMP protomer

PDB-8xq9: 
Structure of the sea urchin spSLC9C1 in state-2 w/ cAMP dimer

PDB-8xqa: 
Structure of the sea urchin spSLC9C1 in state-3 w/ cAMP dimer

EMDB-38617: 
SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab

EMDB-38618: 
SARS-CoV-2 RBD + IMCAS-364 + hACE2

EMDB-38619: 
SARS-CoV-2 RBD + IMCAS-364 (Local Refinement)

EMDB-38620: 
SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2

EMDB-38621: 
SARS-CoV-2 spike + IMCAS-123

EMDB-38823: 
Cryo-EM structure of the 123-316 scDb/PT-RBD complex

PDB-8xse: 
SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab

PDB-8xsf: 
SARS-CoV-2 RBD + IMCAS-364 + hACE2

PDB-8xsi: 
SARS-CoV-2 RBD + IMCAS-364 (Local Refinement)

PDB-8xsj: 
SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2

PDB-8xsl: 
SARS-CoV-2 spike + IMCAS-123

PDB-8y0y: 
Cryo-EM structure of the 123-316 scDb/PT-RBD complex

EMDB-45655: 
Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin
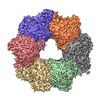
EMDB-36725: 
Acyl-ACP Synthetase structure bound to C10-AMS

PDB-8jyl: 
Acyl-ACP Synthetase structure bound to C10-AMS
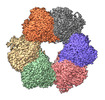
EMDB-36731: 
Acyl-ACP Synthetase structure bound to Decanoyl-AMP

PDB-8jyu: 
Acyl-ACP Synthetase structure bound to Decanoyl-AMP

EMDB-41041: 
Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL

PDB-8t50: 
Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL

EMDB-37105: 
ATP-bound hMRP5 outward-open
EMDB-37554: 
wt-hMRP5 inward-open
EMDB-37555: 
RD-hMRP5-inward open
EMDB-37556: 
ND-hMRP5-inward open
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
