-Search query
-Search result
Showing 1 - 50 of 4,128 items for (author: hong & m)

EMDB entry, No image
EMDB-45623: 
MicroED structure of the C11 cysteine protease clostripain

PDB-9cip: 
MicroED structure of the C11 cysteine protease clostripain

EMDB entry, No image
EMDB-43814: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage

EMDB entry, No image
EMDB-43815: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage

PDB-9ash: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage

PDB-9asi: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage

EMDB entry, No image
EMDB-37139: 
Structure of SARS-CoV Spike protein complexed with antibody PW5-5

EMDB entry, No image
EMDB-37143: 
The local refined map of SARS-CoV-2 XBB Variant Spike protein complexed with antibody PW5-535

EMDB entry, No image
EMDB-37144: 
Trimer state of SARS-CoV Spike protein complexed with antibody PW5-535

EMDB entry, No image
EMDB-37145: 
The local refined map of SARS-CoV Spike protein complexed with antibody PW5-5

EMDB entry, No image
EMDB-37160: 
Monomer state of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5

EMDB entry, No image
EMDB-37161: 
Monomer state of SARS-CoV Spike protein complexed with antibody PW5-535

EMDB entry, No image
EMDB-37162: 
Structure of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570

EMDB entry, No image
EMDB-37163: 
The local refined map of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570

EMDB entry, No image
EMDB-37164: 
State 1 of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5

EMDB entry, No image
EMDB-37165: 
Structure of SARS-CoV-2 XBB Variant Spike protein complexed with broadly neutralizing antibody PW5-535

PDB-8kdm: 
Structure of SARS-CoV Spike protein complexed with antibody PW5-5

PDB-8kdr: 
The local refined map of SARS-CoV-2 XBB Variant Spike protein complexed with antibody PW5-535

PDB-8kds: 
Trimer state of SARS-CoV Spike protein complexed with antibody PW5-535

PDB-8kdt: 
The local refined map of SARS-CoV Spike protein complexed with antibody PW5-5

PDB-8kej: 
Monomer state of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5

PDB-8kek: 
Monomer state of SARS-CoV Spike protein complexed with antibody PW5-535

PDB-8keo: 
Structure of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570

PDB-8kep: 
The local refined map of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570

PDB-8keq: 
State 1 of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5

PDB-8ker: 
Structure of SARS-CoV-2 XBB Variant Spike protein complexed with broadly neutralizing antibody PW5-535
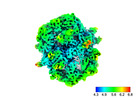
EMDB-45235: 
Subtomogram average (C1) of fatty acid synthase from S.cerevisiae prepared using cryo-plasmaFIB milling

EMDB-37499: 
Cryo-EM structure of CRISPR-Csm effector complex from Mycobacterium canettii

PDB-8wfx: 
Cryo-EM structure of CRISPR-Csm effector complex from Mycobacterium canettii

EMDB-38873: 
cryo-EM structure of Staphylococcus aureus(ATCC 29213) 50S ribosome in complex with MCX-190.

EMDB-38874: 
Cryo-EM structure of Staphylococcus aureus (15B196) 50S ribosome in complex with MCX-190.

EMDB-38875: 
Cryo-EM structure of Staphylococcus aureus 70S ribosome (strain 15B196) in complex with MCX-190.

EMDB-38876: 
cryo-EM structure of Staphylococcus aureus(ATCC 29213) 70S ribosome in complex with MCX-190.

PDB-8y36: 
cryo-EM structure of Staphylococcus aureus(ATCC 29213) 50S ribosome in complex with MCX-190.

PDB-8y37: 
Cryo-EM structure of Staphylococcus aureus (15B196) 50S ribosome in complex with MCX-190.

PDB-8y38: 
Cryo-EM structure of Staphylococcus aureus 70S ribosome (strain 15B196) in complex with MCX-190.

PDB-8y39: 
cryo-EM structure of Staphylococcus aureus(ATCC 29213) 70S ribosome in complex with MCX-190.

EMDB-43046: 
Full length integrin AlphaIIbBeta3 in pre-active state

EMDB-32979: 
Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10)

PDB-7x35: 
Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10)

EMDB-29620: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B)
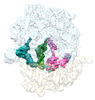
EMDB-29621: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A)
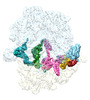
EMDB-29627: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A)
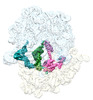
EMDB-29628: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D)

EMDB-29631: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B)
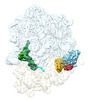
EMDB-29634: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C)

PDB-8fzd: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B)

PDB-8fze: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A)

PDB-8fzg: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A)

PDB-8fzh: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
