+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full length integrin AlphaIIbBeta3 in pre-active state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | integrin / CELL ADHESION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.46 Å | |||||||||
 Authors Authors | Huo T / Wu H / Wang Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Full-length αIIbβ3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture. Authors: Tong Huo / Hongjiang Wu / Zeinab Moussa / Mehmet Sen / Valerie Dalton / Zhao Wang /  Abstract: Integrin αIIbβ3 is the key receptor regulating platelet retraction and accumulation and a proven drug-target for antithrombotic therapies. Here we resolve the cryo-EM structures of the full-length ...Integrin αIIbβ3 is the key receptor regulating platelet retraction and accumulation and a proven drug-target for antithrombotic therapies. Here we resolve the cryo-EM structures of the full-length αIIbβ3, which covers three distinct states along the activation pathway. Firstly, we obtain the αIIbβ3 structure at 3 Å resolution in the inactive state, revealing the overall topology of the heterodimer with the transmembrane (TM) helices and the ligand-binding domain tucked in a specific angle proximity to the TM region. After the addition of a Mn agonist, we resolve two coexisting structures representing two new states between inactive and active state. Our structures show conformational changes of the αIIbβ3 activating trajectory and a unique twisting of the integrin legs, which is required for platelets accumulation. Our structure provides direct structural evidence for how the lower legs are involved in full-length integrin activation mechanisms and offers a new strategy to target the αIIbβ3 lower leg. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43046.map.gz emd_43046.map.gz | 117.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43046-v30.xml emd-43046-v30.xml emd-43046.xml emd-43046.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43046.png emd_43046.png | 24 KB | ||
| Filedesc metadata |  emd-43046.cif.gz emd-43046.cif.gz | 5.4 KB | ||
| Others |  emd_43046_half_map_1.map.gz emd_43046_half_map_1.map.gz emd_43046_half_map_2.map.gz emd_43046_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43046 http://ftp.pdbj.org/pub/emdb/structures/EMD-43046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43046 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43046.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43046.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
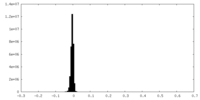
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_43046_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
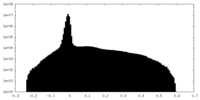
| Density Histograms |
-Half map: #2
| File | emd_43046_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The extracellular domain of integrin AlphaIIbBeta3 in pre-active state
| Entire | Name: The extracellular domain of integrin AlphaIIbBeta3 in pre-active state |
|---|---|
| Components |
|
-Supramolecule #1: The extracellular domain of integrin AlphaIIbBeta3 in pre-active state
| Supramolecule | Name: The extracellular domain of integrin AlphaIIbBeta3 in pre-active state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Integrin alpha-IIb
| Macromolecule | Name: Integrin alpha-IIb / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARALCPLQA LWLLEWVLLL LGPCAAPPAW ALNLDPVQLT FYAGPNGSQF GFSLDFHKDS HGRVAIVVG APRTLGPSQE ETGGVFLCPW RAEGGQCPSL LFDLRDETRN VGSQTLQTFK A RQGLGASV VSWSDVIVAC APWQHWNVLE KTEEAEKTPV GSCFLAQPES ...String: MARALCPLQA LWLLEWVLLL LGPCAAPPAW ALNLDPVQLT FYAGPNGSQF GFSLDFHKDS HGRVAIVVG APRTLGPSQE ETGGVFLCPW RAEGGQCPSL LFDLRDETRN VGSQTLQTFK A RQGLGASV VSWSDVIVAC APWQHWNVLE KTEEAEKTPV GSCFLAQPES GRRAEYSPCR GN TLSRIYV ENDFSWDKRY CEAGFSSVVT QAGELVLGAP GGYYFLGLLA QAPVADIFSS YRP GILLWH VSSQSLSFDS SNPEYFDGYW GYSVAVGEFD GDLNTTEYVV GAPTWSWTLG AVEI LDSYY QRLHRLRGEQ MASYFGHSVA VTDVNGDGRH DLLVGAPLYM ESRADRKLAE VGRVY LFLQ PRGPHALGAP SLLLTGTQLY GRFGSAIAPL GDLDRDGYND IAVAAPYGGP SGRGQV LVF LGQSEGLRSR PSQVLDSPFP TGSAFGFSLR GAVDIDDNGY PDLIVGAYGA NQVAVYR AQ PVVKASVQLL VQDSLNPAVK SCVLPQTKTP VSCFNIQMCV GATGHNIPQK LSLNAELQ L DRQKPRQGRR VLLLGSQQAG TTLNLDLGGK HSPICHTTMA FLRDEADFRD KLSPIVLSL NVSLPPTEAG MAPAVVLHGD THVQEQTRIV LDCGEDDVCV PQLQLTASVT GSPLLVGADN VLELQMDAA NEGEGAYEAE LAVHLPQGAH YMRALSNVEG FERLICNQKK ENETRVVLCE L GNPMKKNA QIGIAMLVSV GNLEEAGESV SFQLQIRSKN SQNPNSKIVL LDVPVRAEAQ VE LRGNSFP ASLVVAAEEG EREQNSLDSW GPKVEHTYEL HNNGPGTVNG LHLSIHLPGQ SQP SDLLYI LDIQPQGGLQ CFPQPPVNPL KVDWGLPIPS PSPIHPAHHK RDRRQIFLPE PEQP SRLQD PVLVSCDSAP CTVVQCDLQE MARGQRAMVT VLAFLWLPSL YQRPLDQFVL QSHAW FNVS SLPYAVPPLS LPRGEAQVWT QLLRALEERA IPIWWVLVGV LGGLLLLTIL VLAMWK VGF FKRNRPPLEE DDEEGE |
-Macromolecule #2: Integrin beta-3
| Macromolecule | Name: Integrin beta-3 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRARPRPRPL WATVLALGAL AGVGVGGPNI CTTRGVSSCQ QCLAVSPMCA WCSDEALPLG SPRCDLKEN LLKDNCAPES IEFPVSEARV LEDRPLSDKG SGDSSQVTQV SPQRIALRLR P DDSKNFSI QVRQVEDYPV DIYYLMDLSY SMKDDLWSIQ NLGTKLATQM ...String: MRARPRPRPL WATVLALGAL AGVGVGGPNI CTTRGVSSCQ QCLAVSPMCA WCSDEALPLG SPRCDLKEN LLKDNCAPES IEFPVSEARV LEDRPLSDKG SGDSSQVTQV SPQRIALRLR P DDSKNFSI QVRQVEDYPV DIYYLMDLSY SMKDDLWSIQ NLGTKLATQM RKLTSNLRIG FG AFVDKPV SPYMYISPPE ALENPCYDMK TTCLPMFGYK HVLTLTDQVT RFNEEVKKQS VSR NRDAPE GGFDAIMQAT VCDEKIGWRN DASHLLVFTT DAKTHIALDG RLAGIVQPND GQCH VGSDN HYSASTTMDY PSLGLMTEKL SQKNINLIFA VTENVVNLYQ NYSELIPGTT VGVLS MDSS NVLQLIVDAY GKIRSKVELE VRDLPEELSL SFNATCLNNE VIPGLKSCMG LKIGDT VSF SIEAKVRGCP QEKEKSFTIK PVGFKDSLIV QVTFDCDCAC QAQAEPNSHR CNNGNGT FE CGVCRCGPGW LGSQCECSEE DYRPSQQDEC SPREGQPVCS QRGECLCGQC VCHSSDFG K ITGKYCECDD FSCVRYKGEM CSGHGQCSCG DCLCDSDWTG YYCNCTTRTD TCMSSNGLL CSGRGKCECG SCVCIQPGSY GDTCEKCPTC PDACTFKKEC VECKKFDRGA LHDENTCNRY CRDEIESVK ELKDTGKDAV NCTYKNEDDC VVRFQYYEDS SGKSILYVVE EPECPKGPDI L VVLLSVMG AILLIGLAAL LIWKLLITIH DRKEFAKFEE ERARAKWDTA NNPLYKEATS TF TNITYRG T |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.46 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 175020 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)