-Search query
-Search result
Showing 1 - 50 of 108 items for (author: gumbart & j)

EMDB-49494: 
Thermothelomyces thermophilus SAM complex closed conformation
Method: single particle / : Diederichs K, Botos I, Buchanan SK

EMDB-49495: 
Thermothelomyces thermophilus SAM complex open conformation
Method: single particle / : Diederichs K, Botos I, Buchanan SK

EMDB-49496: 
Thermothelomyces thermophilus SAM complex bound to darobactin A
Method: single particle / : Diederichs K, Botos I, Buchanan SK

PDB-9nk6: 
Thermothelomyces thermophilus SAM complex closed conformation
Method: single particle / : Diederichs K, Botos I, Buchanan SK

PDB-9nk7: 
Thermothelomyces thermophilus SAM complex open conformation
Method: single particle / : Diederichs K, Botos I, Buchanan SK

PDB-9nk8: 
Thermothelomyces thermophilus SAM complex bound to darobactin A
Method: single particle / : Diederichs K, Botos I, Buchanan SK

EMDB-45754: 
The cryo-EM structure of BamACDE complex from Neisseria gonorrhoeae
Method: single particle / : Billings EM, Noinaj N

EMDB-45755: 
The cryo-EM structure of BamADE subcomplex from Neisseria gonorrhoeae
Method: single particle / : Billings EM, Noinaj N

EMDB-45756: 
The cryo-EM structure of BamAD subcomplex from Neisseria gonorrhoeae
Method: single particle / : Billings EM, Noinaj N

PDB-9cmw: 
The cryo-EM structure of BamACDE complex from Neisseria gonorrhoeae
Method: single particle / : Billings EM, Noinaj N

PDB-9cn0: 
The cryo-EM structure of BamADE subcomplex from Neisseria gonorrhoeae
Method: single particle / : Billings EM, Noinaj N

PDB-9cn1: 
The cryo-EM structure of BamAD subcomplex from Neisseria gonorrhoeae
Method: single particle / : Billings EM, Noinaj N

EMDB-40682: 
The cryo-EM structure of the EcBAM/EspP(beta1-12) complex
Method: single particle / : Wu R, Noinaj N

EMDB-40700: 
The cryo-EM structure of the EcBAM/EspP(beta8-12) complex
Method: single particle / : Wu R, Noinaj N

EMDB-40701: 
The cryo-EM structure of the EcBAM/EspP(beta7-12) complex
Method: single particle / : Wu R, Noinaj N

EMDB-41508: 
Cryo-EM structure of E3 ubiquitin ligase Doa10 from Saccharomyces cerevisiae
Method: single particle / : Park E, Itskanov SI

PDB-8tqm: 
Cryo-EM structure of E3 ubiquitin ligase Doa10 from Saccharomyces cerevisiae
Method: single particle / : Park E, Itskanov SI

EMDB-27825: 
Cryo-EM structure of the endogenous core TIM23 complex from S. cerevisiae
Method: single particle / : Sim SI, Park E

EMDB-40346: 
Cryo-EM structure of the core TIM23 complex from S. cerevisiae
Method: single particle / : Sim SI, Park E

PDB-8e1m: 
Cryo-EM structure of the endogenous core TIM23 complex from S. cerevisiae
Method: single particle / : Sim SI, Park E

PDB-8scx: 
Cryo-EM structure of the core TIM23 complex from S. cerevisiae
Method: single particle / : Sim SI, Park E

EMDB-24224: 
Combined 3D structure of the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echeverria I, Fernandez-Martinez J, Nudelman I

EMDB-24225: 
Low resolution, multi-part 3D structure of the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echeverria I, Fernandez-Martinez J, Nudelman I

EMDB-24231: 
Double nuclear outer ring from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP

EMDB-24232: 
Inner ring spoke from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP

EMDB-24258: 
Structure of the in situ yeast NPC
Method: subtomogram averaging / : Villa E, Singh D
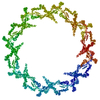
PDB-7n84: 
Double nuclear outer ring from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echeverria I, Fernandez-Martinez J, Nudelman I
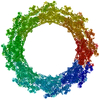
PDB-7n85: 
Inner ring spoke from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echeverria I, Fernandez-Martinez J, Nudelman I

PDB-7n9f: 
Structure of the in situ yeast NPC
Method: subtomogram averaging / : Villa E, Singh D, Ludtke SJ, Akey CW, Rout MP, Echeverria I, Suslov S

EMDB-24473: 
Structure of a BAM/EspP(beta9-12) hybrid-barrel intermediate
Method: single particle / : Wu RR, Noinaj N

EMDB-24474: 
Structure of a BAM in MSP1E3D1 nanodiscs at 4 Angstrom resolution
Method: single particle / : Wu RR, Noinaj N

EMDB-24475: 
Structure of BAM in MSP1E3D1 nanodiscs prepared from E. coli outer membranes
Method: single particle / : Wu RR, Noinaj N

EMDB-24476: 
The structure of BAM in MSP1D1 nanodiscs
Method: single particle / : Wu RR, Noinaj N

EMDB-24477: 
The structure of BAM in MSP2N2 nanodiscs
Method: single particle / : Wu RR, Noinaj N

EMDB-24478: 
The structure of BAM in MSP1E3D1 at 6.9 Angstrom resolution
Method: single particle / : Wu RR, Noinaj N

EMDB-24481: 
The structure of BAM in complex with EspP at 7 Angstrom resolution
Method: single particle / : Wu RR, Noinaj N

PDB-7ri4: 
Structure of a BAM/EspP(beta9-12) hybrid-barrel intermediate
Method: single particle / : Wu RR, Noinaj N

PDB-7ri5: 
Structure of a BAM in MSP1E3D1 nanodiscs at 4 Angstrom resolution
Method: single particle / : Wu RR, Noinaj N

PDB-7ri6: 
Structure of BAM in MSP1E3D1 nanodiscs prepared from E. coli outer membranes
Method: single particle / : Wu RR, Noinaj N

PDB-7ri9: 
The structure of BAM in MSP1E3D1 at 6.9 Angstrom resolution
Method: single particle / : Wu RR, Noinaj N

PDB-7rj5: 
The structure of BAM in complex with EspP at 7 Angstrom resolution
Method: single particle / : Wu RR, Noinaj N

EMDB-22770: 
Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class without Sec62
Method: single particle / : Itskanov S, Park E

EMDB-22773: 
Cryo-EM structure of the Sec complex from T. lanuginosus, wild-type, class without Sec62
Method: single particle / : Itskanov S, Park E

EMDB-22776: 
Cryo-EM structure of the Sec complex from T. lanuginosus lacking Sec62
Method: single particle / : Itskanov S, Park E

EMDB-22777: 
Cryo-EM structure of the Sec complex from T. lanuginosus, Sec62 anchor domain mutant (delta anchor)
Method: single particle / : Itskanov S, Park E

EMDB-22778: 
Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class without Sec62
Method: single particle / : Itskanov S, Park E

EMDB-22785: 
Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, consensus map
Method: single particle / : Itskanov S, Park E

EMDB-22786: 
Cryo-EM structure of the Sec complex from T. lanuginosus, wild-type, consensus map of classes with Sec62
Method: single particle / : Itskanov S, Park E
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
