+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24258 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the in situ yeast NPC | ||||||||||||||||||||||||
 Map data Map data | full in situ NPC map recombined from 90 degree wedge focused alignment | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | NPC / nucleocytoplasmic transport / TRANSLOCASE | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to spindle checkpoint signaling / nuclear pore linkers / Regulation of Glucokinase by Glucokinase Regulatory Protein / regulation of protein desumoylation / mRNA export from nucleus in response to heat stress / peroxisomal importomer complex / positive regulation of ER to Golgi vesicle-mediated transport / Seh1-associated complex / chromosome, subtelomeric region / nuclear pore inner ring ...response to spindle checkpoint signaling / nuclear pore linkers / Regulation of Glucokinase by Glucokinase Regulatory Protein / regulation of protein desumoylation / mRNA export from nucleus in response to heat stress / peroxisomal importomer complex / positive regulation of ER to Golgi vesicle-mediated transport / Seh1-associated complex / chromosome, subtelomeric region / nuclear pore inner ring / COPII-coated vesicle budding / nuclear pore localization / protein exit from endoplasmic reticulum / adenyl-nucleotide exchange factor activity / nuclear pore central transport channel / transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery / regulation of nucleocytoplasmic transport / COPII-mediated vesicle transport / nuclear pore complex assembly / regulation of TORC1 signaling / telomere tethering at nuclear periphery / nuclear pore outer ring / protein localization to nuclear inner membrane / nuclear pore organization / nuclear migration along microtubule / positive regulation of protein exit from endoplasmic reticulum / nuclear pore cytoplasmic filaments / : / COPII vesicle coat / Regulation of HSF1-mediated heat shock response / establishment of mitotic spindle localization / post-transcriptional tethering of RNA polymerase II gene DNA at nuclear periphery / nuclear pore nuclear basket / protein localization to kinetochore / tRNA export from nucleus / SUMOylation of SUMOylation proteins / structural constituent of nuclear pore / SUMOylation of RNA binding proteins / RNA export from nucleus / SUMOylation of chromatin organization proteins / silent mating-type cassette heterochromatin formation / NLS-bearing protein import into nucleus / vacuolar membrane / nucleocytoplasmic transport / nuclear localization sequence binding / cytoplasmic dynein complex / poly(A)+ mRNA export from nucleus / regulation of mitotic nuclear division / dynein intermediate chain binding / establishment of mitotic spindle orientation / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / ribosomal large subunit export from nucleus / positive regulation of TOR signaling / mRNA transport / nuclear pore / subtelomeric heterochromatin formation / mRNA export from nucleus / cytoplasmic microtubule / ERAD pathway / ribosomal small subunit export from nucleus / Neutrophil degranulation / positive regulation of TORC1 signaling / nuclear periphery / protein export from nucleus / cellular response to amino acid starvation / cell periphery / chromosome segregation / promoter-specific chromatin binding / molecular condensate scaffold activity / phospholipid binding / protein import into nucleus / transcription corepressor activity / nuclear envelope / heterochromatin formation / double-strand break repair / single-stranded DNA binding / protein transport / cellular response to heat / nuclear membrane / amyloid fibril formation / chromosome, telomeric region / hydrolase activity / cell division / chromatin binding / endoplasmic reticulum membrane / positive regulation of DNA-templated transcription / protein-containing complex binding / structural molecule activity / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / positive regulation of transcription by RNA polymerase II / DNA binding / RNA binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
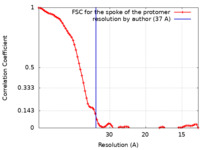
| Method | subtomogram averaging / cryo EM / Resolution: 37.0 Å | ||||||||||||||||||||||||
 Authors Authors | Villa E / Singh D | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Authors: Christopher W Akey / Digvijay Singh / Christna Ouch / Ignacia Echeverria / Ilona Nudelman / Joseph M Varberg / Zulin Yu / Fei Fang / Yi Shi / Junjie Wang / Daniel Salzberg / Kangkang Song / ...Authors: Christopher W Akey / Digvijay Singh / Christna Ouch / Ignacia Echeverria / Ilona Nudelman / Joseph M Varberg / Zulin Yu / Fei Fang / Yi Shi / Junjie Wang / Daniel Salzberg / Kangkang Song / Chen Xu / James C Gumbart / Sergey Suslov / Jay Unruh / Sue L Jaspersen / Brian T Chait / Andrej Sali / Javier Fernandez-Martinez / Steven J Ludtke / Elizabeth Villa / Michael P Rout /  Abstract: Nuclear pore complexes (NPCs) mediate the nucleocytoplasmic transport of macromolecules. Here we provide a structure of the isolated yeast NPC in which the inner ring is resolved by cryo-EM at sub- ...Nuclear pore complexes (NPCs) mediate the nucleocytoplasmic transport of macromolecules. Here we provide a structure of the isolated yeast NPC in which the inner ring is resolved by cryo-EM at sub-nanometer resolution to show how flexible connectors tie together different structural and functional layers. These connectors may be targets for phosphorylation and regulated disassembly in cells with an open mitosis. Moreover, some nucleoporin pairs and transport factors have similar interaction motifs, which suggests an evolutionary and mechanistic link between assembly and transport. We provide evidence for three major NPC variants that may foreshadow functional specializations at the nuclear periphery. Cryo-electron tomography extended these studies, providing a model of the in situ NPC with a radially expanded inner ring. Our comprehensive model reveals features of the nuclear basket and central transporter, suggests a role for the lumenal Pom152 ring in restricting dilation, and highlights structural plasticity that may be required for transport. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24258.map.gz emd_24258.map.gz | 76.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24258-v30.xml emd-24258-v30.xml emd-24258.xml emd-24258.xml | 68.6 KB 68.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24258_fsc.xml emd_24258_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_24258.png emd_24258.png | 197.2 KB | ||
| Masks |  emd_24258_msk_1.map emd_24258_msk_1.map emd_24258_msk_2.map emd_24258_msk_2.map emd_24258_msk_3.map emd_24258_msk_3.map | 34.3 MB 34.3 MB 34.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24258.cif.gz emd-24258.cif.gz | 18.9 KB | ||
| Others |  emd_24258_additional_1.map.gz emd_24258_additional_1.map.gz emd_24258_additional_2.map.gz emd_24258_additional_2.map.gz emd_24258_additional_3.map.gz emd_24258_additional_3.map.gz emd_24258_additional_4.map.gz emd_24258_additional_4.map.gz emd_24258_additional_5.map.gz emd_24258_additional_5.map.gz emd_24258_additional_6.map.gz emd_24258_additional_6.map.gz emd_24258_additional_7.map.gz emd_24258_additional_7.map.gz emd_24258_half_map_1.map.gz emd_24258_half_map_1.map.gz emd_24258_half_map_2.map.gz emd_24258_half_map_2.map.gz | 111 MB 111.7 MB 113.8 MB 111.9 MB 111.8 MB 111.2 MB 112.2 MB 12.7 MB 12.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24258 http://ftp.pdbj.org/pub/emdb/structures/EMD-24258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24258 | HTTPS FTP |
-Related structure data
| Related structure data |  7n9fMC  7n84C  7n85C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24258.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24258.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full in situ NPC map recombined from 90 degree wedge focused alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.74 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
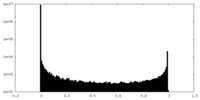
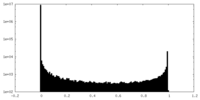
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Mask #2
+Mask #3
+Additional map: zoned 3D map of Nsp1-FG-connections
+Additional map: zoned 3D map of inner ring of spokes
+Additional map: full in situ NPC map recombined from 90...
+Additional map: zoned 3D map of the N-ring
+Additional map: zoned 3D map of C-ring plus Nup82 complex with 3 Dyn 2 dimers
+Additional map: zoned 3D map of Pom152 ring
+Additional map: difference map for pore membrane
+Half map: half map
+Half map: half map
- Sample components
Sample components
+Entire : yeast NPC
+Supramolecule #1: yeast NPC
+Macromolecule #1: Nucleoporin NUP170
+Macromolecule #2: Nucleoporin NUP157
+Macromolecule #3: orphans bound to Nup192 NTD
+Macromolecule #4: Nucleoporin NSP1
+Macromolecule #5: Nucleoporin NUP57
+Macromolecule #6: Nucleoporin NUP49/NSP49
+Macromolecule #7: Nucleoporin NUP192
+Macromolecule #8: Nucleoporin NUP188
+Macromolecule #9: Nucleoporin NIC96
+Macromolecule #10: Nucleoporin NUP53
+Macromolecule #11: Nucleoporin ASM4
+Macromolecule #12: Nucleoporin NUP82
+Macromolecule #13: Nucleoporin NUP159
+Macromolecule #14: Nucleoporin NUP120
+Macromolecule #15: Nucleoporin NUP85
+Macromolecule #16: Nucleoporin 145c
+Macromolecule #17: Protein transport protein SEC13
+Macromolecule #18: Nucleoporin SEH1
+Macromolecule #19: Nucleoporin NUP84
+Macromolecule #20: Nucleoporin NUP133
+Macromolecule #21: Dynein light chain 1, cytoplasmic
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE-PROPANE Details: A custom-built vitrification device (Max Planck Institute for Biochemistry, Munich). |
| Details | W303 yeast cells were harvested during log-phase growth and diluted to an 0.8 x 107 cells/mL in YPD media. Five uL of this diluted sample was applied to glow-discharged 200-mesh, Quantafoil R2/1 grids (Electron Microscopy Sciences), excess media was manually blotted from the grid back side (opposite to the carbon substrate where the cells were deposited) and the grid was plunge frozen in a liquid ethane-propane mixture (50/50 volume, Airgas) then cryogenic, FIB milling was performed in an Aquilos DualBeam (Thermo Fisher Scientific) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Details | Tilt series collection: dose-symmetric & bi-directional |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3708 pixel / Average exposure time: 2.0 sec. / Average electron dose: 4.5 e/Å2 Details: Images were collected in movie-mode at ~8 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 6.0 µm / Calibrated defocus min: 2.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 42000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Manual docking and limited molecular dynamics flexible fitting |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: cross-correlation |
| Output model |  PDB-7n9f: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)