-Search query
-Search result
Showing 1 - 50 of 203 items for (author: friedrich & t)

EMDB-19212: 
in situ subtomogram average of MEF cell ribosome in the decoding Z state

EMDB-19213: 
in situ subtomogram average of MEF cell ribosome in the PRE+ Z state

EMDB-19214: 
in situ subtomogram average of MEF cell ribosome in a PRE+ state

EMDB-19215: 
in situ subtomogram average of MEF cell ribosome in a different PRE+ state

EMDB-19216: 
in situ subtomogram average of MEF cell ribosome in the classical PRE state

EMDB-19217: 
in situ subtomogram average of MEF cell ribosome in the rotated 2 state

EMDB-19218: 
in situ subtomogram average of MEF cell ribosome in the rotated 2 + state

EMDB-19219: 
in situ subtomogram average of MEF cell ribosome in a translocation intermediate POSTi state

EMDB-19220: 
in situ subtomogram average of MEF cell ribosome in the POST state

EMDB-19221: 
in situ subtomogram average of low dose anisomycin treated MEF cell ribosome in the OFF-P state

EMDB-19222: 
in situ subtomogram average of MEF cell pre-60S ribosome in the state B

EMDB-19223: 
in situ subtomogram average of MEF cell idle 60S ribosome complex

EMDB-19224: 
in situ subtomogram average of MEF cell ribosome associated quality control complex

EMDB-19225: 
in situ subtomogram average of MEF cell non-empty 60S ribosome complex

EMDB-19226: 
in situ subtomogram average of MEF cell 40S ribosome

EMDB-19227: 
in situ subtomogram average of MEF cell 48S initiation complexes

EMDB-19228: 
in situ subtomogram average of high dose anisomycin treated MEF cell ribosome in PRE+ Z state

EMDB-19229: 
in situ subtomogram average of an aberrant 40S initiation complex in low dose anisomycin (20 min) treated MEF cell

EMDB-19230: 
n situ subtomogram average of aberrant initiation complex in arsenite treated MEF cells

EMDB-19231: 
in situ subtomogram average of 43S initiation complex in low dose anisomycin treated MEF cells

EMDB-19232: 
in situ subtomogram average of a subclass of 43S initiation complex in low dose anisomycin treated MEF cells

EMDB-19233: 
in situ subtomogram average of an aberrant 40S initiation complex in low dose anisomycin treated MEF cells

EMDB-19234: 
in situ subtomogram average of an aberrant 40S initiation complex in low dose anisomycin treated MEF cells

EMDB-19235: 
in situ subtomogram average of decoding-like stalled ribosome in low dose anisomycin treated MEF cells

EMDB-19236: 
in situ subtomogram average of PRE-like stalled ribosome in low dose anisomycin treated MEF cells

EMDB-19237: 
in situ subtomogram average of rotated 2 collided ribosome in low dose anisomycin treated MEF cells

EMDB-19238: 
in situ subtomogram average of decoding-like collided ribosome in low dose anisomycin treated MEF cells

EMDB-19239: 
in situ subtomogram average of POSTi-like middle ribosome in helical polysomes in low dose anisomycin treated MEF cells

EMDB-19240: 
in situ subtomogram average of GCN1-bound stalled ribosome in low dose anisomycin treated MEF cells

EMDB-19242: 
in situ subtomogram average of GCN1-bound collided ribosome in low dose anisomycin treated MEF cells

EMDB-19211: 
in situ subtomogram average of MEF cell ribosomes in the decoding E state

EMDB-19035: 
Composite map of the Emiliania huxleyi virus 201 (EhV-201) symmetry expanded from cryo-EM structure of virion vertex 120 nm in diameter.

EMDB-19036: 
Cryo-EM structure of the Emiliania huxleyi virus 201 (EhV-201) virion vertex with a diameter of 50 nm and a mask applied on the capsid layer.

PDB-8rbs: 
Emiliania huxleyi virus 201 (EhV-201) asymmetrical unit of capsid proteins predicted by AlphaFold2 fitted into the cryo-EM density of EhV-201 virion composite map.

PDB-8rbt: 
Emiliania huxleyi virus 201 (EhV-201) capsid proteins predicted by AlphaFold2 fitted into a cryo-EM density map of the EhV-201 virion capsid.

EMDB-17823: 
Tomogram of the Emiliania huxleyi virus 201 (EhV-201) purified particles used for subtomogram averaging.

EMDB-17649: 
Cryo-EM structure of the Emiliania huxleyi virus 201 (EhV-201) virion vertex with a diameter of 50 nm.

EMDB-17650: 
Cryo-EM structure of the Emiliania huxleyi virus 201 (EhV-201) virion vertex with a diameter of 50 nm.

EMDB-17651: 
Cryo-EM structure of the Emiliania huxleyi virus 201 (EhV-201) virion vertex with a diameter of 120 nm.
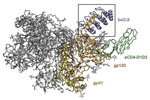
EMDB-26157: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9

PDB-7txd: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9

EMDB-15532: 
Plasmodium falciparum sporozoite subpellicular microtubule with interrupted luminal helix determined in situ

EMDB-15534: 
Plasmodium falciparum gametocyte subpellicular microtubule with 13 protofilaments determined in situ

EMDB-15535: 
Plasmodium falciparum gametocyte subpellicular microtubule with 14 protofilaments determined in situ

EMDB-15536: 
Plasmodium falciparum gametocyte subpellicular microtubule with 15 protofilaments determined in situ

EMDB-15537: 
Plasmodium falciparum gametocyte subpellicular microtubule with 16 protofilaments determined in situ

EMDB-15538: 
Plasmodium falciparum gametocyte subpellicular microtubule with 17 protofilaments determined in situ

EMDB-15539: 
Plasmodium falciparum gametocyte subpellicular microtubule with 18 protofilaments determined in situ
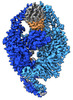
EMDB-15417: 
human MutSalpha (MSH2/MSH6) binding to DNA with a GT mismatch
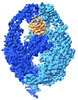
EMDB-15519: 
human MutSalpha (MSH2/MSH6) on DNA containing a GT mismatch in the presence of ADP
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
