-Search query
-Search result
Showing 1 - 50 of 97 items for (author: fremont & dh)

EMDB-42187: 
Eastern equine encephalitis virus (PE-6) VLP (icosahedral)

EMDB-42189: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (icosahedral)

EMDB-42191: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (icosahedral)

EMDB-42193: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (icosahedral)

EMDB-42194: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (asymmetric unit)

EMDB-42195: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (icosahedral)

EMDB-42196: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (asymmetric unit)

EMDB-42197: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (icosahedral)

EMDB-42198: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (asymmetric unit)

EMDB-42199: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (icosahedral)

EMDB-42200: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (asymmetric unit)

EMDB-42201: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(3-8) (icosahedral)

EMDB-42202: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (icosahedral)

EMDB-42203: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (asymmetric unit)

EMDB-42212: 
Eastern equine encephalitis virus (FL93-939) VLP in complex with VLDLR LA(1-6) (icosahedral)

EMDB-42188: 
Eastern equine encephalitis virus (PE-6) VLP (asymmetric unit)

EMDB-42190: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (asymmetric unit)

EMDB-42192: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (asymmetric unit)

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP

EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor

EMDB-28644: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor

EMDB-27763: 
Chikungunya VLP in complex with neutralizing Fab 506.A08 (asymmetric unit)

EMDB-27764: 
Chikungunya VLP in complex with neutralizing Fab 506.A08 (icosahedral reconstruction)

EMDB-27765: 
Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmetric unit)

EMDB-27766: 
Chikungunya VLP in complex with neutralizing Fab 506.C01 (icosahedral reconstruction)

EMDB-27767: 
Chikungunya VLP in complex with neutralizing Fab CHK-265 (asymmetric unit)

EMDB-27768: 
Chikungunya VLP in complex with neutralizing Fab CHK-265 (icosahedral reconstruction)

EMDB-25487: 
SARS-CoV-2 Spike NTD in complex with neutralizing Fab SARS2-57 (local refinement)

EMDB-25488: 
SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-57 (three down conformation)

EMDB-24394: 
Unsharpened electron microscopy map of Venezuelan equine encephalitis VLP in complex with Domains 1 and 2 of LDLRAD3 receptor

EMDB-24116: 
CryoEM structure of Venezuelan equine encephalitis virus VLP in complex with the LDLRAD3 receptor

EMDB-24117: 
CryoEM structure of Venezuelan equine encephalitis virus VLP

EMDB-22748: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation)

EMDB-22749: 
SARS-CoV-2 Spike RBD in complex with neutralizing Fab 2B04 (local refinement)

EMDB-22750: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2H04 (three down conformation)

EMDB-22751: 
SARS-CoV-2 Spike RBD in complex with neutralizing Fab 2H04 (local refinement)

EMDB-22752: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (two up, one down conformation)

EMDB-22753: 
SARS-CoV-2 Spike in complex with neutralizing Fab 2H04 (one up, two down conformation)

EMDB-23898: 
SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-38 (three down conformation)

EMDB-23899: 
SARS-CoV-2 Spike RBD in complex with neutralizing Fab SARS2-38 (local refinement)
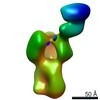
EMDB-22551: 
Polyclonal immune complex of Fab binding the receptor binding site region of H5 HA from serum of subject 36 at day 28

EMDB-22552: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 36 at day 500

EMDB-22553: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 7

EMDB-22554: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 7

EMDB-22555: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 21

EMDB-22556: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 28

EMDB-22557: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 28
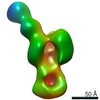
EMDB-22558: 
Polyclonal immune complex of Fab binding the receptor binding site region of H5 HA from serum of subject 43 at day 28

EMDB-22559: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 100
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
